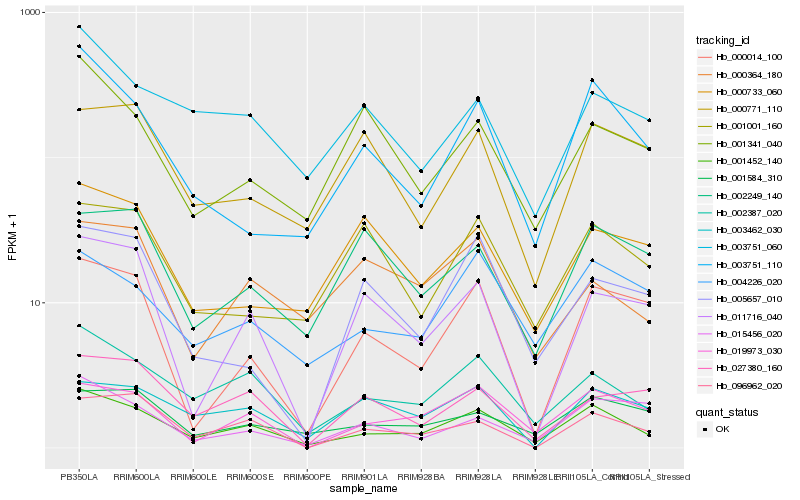
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001452_140 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 2 |
Hb_002387_020 |
0.1071256285 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 3 |
Hb_019973_030 |
0.1574705516 |
- |
- |
- |
| 4 |
Hb_003751_110 |
0.1576705961 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_004226_020 |
0.163552825 |
- |
- |
- |
| 6 |
Hb_001584_310 |
0.1744321416 |
- |
- |
hypothetical protein POPTR_0013s00630g [Populus trichocarpa] |
| 7 |
Hb_096962_020 |
0.1770206526 |
- |
- |
- |
| 8 |
Hb_003751_060 |
0.1800408444 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 9 |
Hb_000014_100 |
0.1858792444 |
- |
- |
- |
| 10 |
Hb_011716_040 |
0.1941194326 |
- |
- |
- |
| 11 |
Hb_003462_030 |
0.1945669034 |
- |
- |
hypothetical protein UMN_5210_01, partial [Pinus radiata] |
| 12 |
Hb_027380_160 |
0.1969885287 |
- |
- |
- |
| 13 |
Hb_015456_020 |
0.1971042348 |
- |
- |
- |
| 14 |
Hb_001341_040 |
0.1972338732 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 15 |
Hb_000364_180 |
0.1972750827 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130 [Jatropha curcas] |
| 16 |
Hb_002249_140 |
0.2034647362 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 17 |
Hb_001001_160 |
0.2042534402 |
- |
- |
Phosphatidic acid phosphohydrolase 2 isoform 1 [Theobroma cacao] |
| 18 |
Hb_000771_110 |
0.2065674072 |
- |
- |
protein phosphatase 2C [Hevea brasiliensis] |
| 19 |
Hb_005657_010 |
0.20742423 |
- |
- |
- |
| 20 |
Hb_000733_060 |
0.2076073011 |
- |
- |
PREDICTED: protein GUCD1 isoform X1 [Jatropha curcas] |