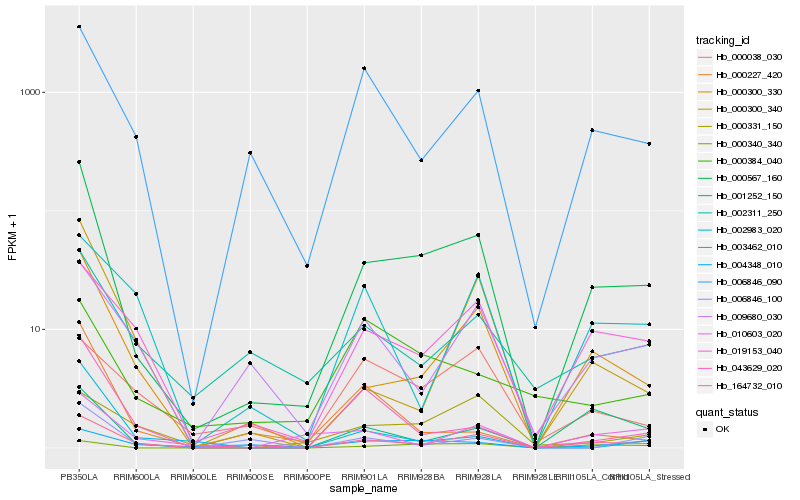
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_160 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 2 |
Hb_164732_010 |
0.1639041559 |
- |
- |
- |
| 3 |
Hb_000300_330 |
0.1879314657 |
- |
- |
- |
| 4 |
Hb_019153_040 |
0.2396295152 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_006846_090 |
0.2449178132 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |
| 6 |
Hb_000340_340 |
0.260799572 |
- |
- |
hypothetical protein POPTR_0014s13260g [Populus trichocarpa] |
| 7 |
Hb_043629_020 |
0.2681260922 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 8 |
Hb_002311_250 |
0.270745961 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 9 |
Hb_009680_030 |
0.2713248733 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 10 |
Hb_000300_340 |
0.2738176213 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001252_150 |
0.2756494924 |
- |
- |
- |
| 12 |
Hb_004348_010 |
0.2787118896 |
- |
- |
hypothetical protein POPTR_0077s002302g, partial [Populus trichocarpa] |
| 13 |
Hb_010603_020 |
0.2885630734 |
- |
- |
- |
| 14 |
Hb_000227_420 |
0.2936521792 |
- |
- |
PREDICTED: thermospermine synthase ACAULIS5-like [Jatropha curcas] |
| 15 |
Hb_006846_100 |
0.298197882 |
- |
- |
hypothetical protein POPTR_0012s11270g [Populus trichocarpa] |
| 16 |
Hb_003462_010 |
0.2986808014 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_000331_150 |
0.3019531443 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_002983_020 |
0.3077409091 |
- |
- |
PREDICTED: probable disease resistance protein At5g63020 [Populus euphratica] |
| 19 |
Hb_000384_040 |
0.3085333484 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 20 |
Hb_000038_030 |
0.3102470333 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |