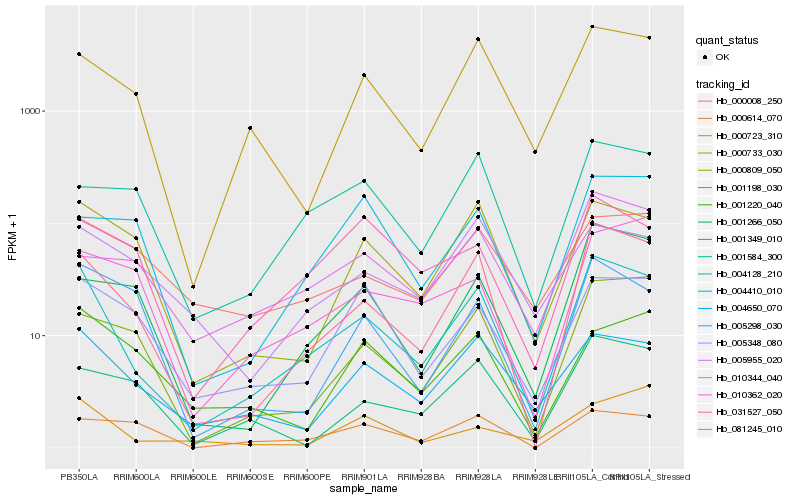
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_210 |
0.0 |
- |
- |
PREDICTED: protein ALUMINUM SENSITIVE 3 [Jatropha curcas] |
| 2 |
Hb_081245_010 |
0.1203789208 |
- |
- |
Caffeic acid 3-O-methyltransferase [Morus notabilis] |
| 3 |
Hb_004650_070 |
0.1695693135 |
- |
- |
- |
| 4 |
Hb_000733_030 |
0.1849579768 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 5 |
Hb_000809_050 |
0.1918204464 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639663 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001198_030 |
0.1974145378 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 7 |
Hb_005348_080 |
0.2074274157 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |
| 8 |
Hb_001266_050 |
0.208644714 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like isoform X2 [Elaeis guineensis] |
| 9 |
Hb_031527_050 |
0.2102154119 |
- |
- |
PREDICTED: universal stress protein A-like protein [Populus euphratica] |
| 10 |
Hb_001349_010 |
0.2115672631 |
- |
- |
- |
| 11 |
Hb_001584_300 |
0.2128866627 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 12 |
Hb_005298_030 |
0.2148909278 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 13 |
Hb_010344_040 |
0.2151296497 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 14 |
Hb_000008_250 |
0.2200910971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005955_020 |
0.2203193327 |
- |
- |
ring finger containing protein, putative [Ricinus communis] |
| 16 |
Hb_000614_070 |
0.2208497807 |
- |
- |
- |
| 17 |
Hb_000723_310 |
0.2238539368 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_010362_020 |
0.2241545897 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 38-like [Jatropha curcas] |
| 19 |
Hb_001220_040 |
0.2244145687 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004410_010 |
0.2280691787 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |