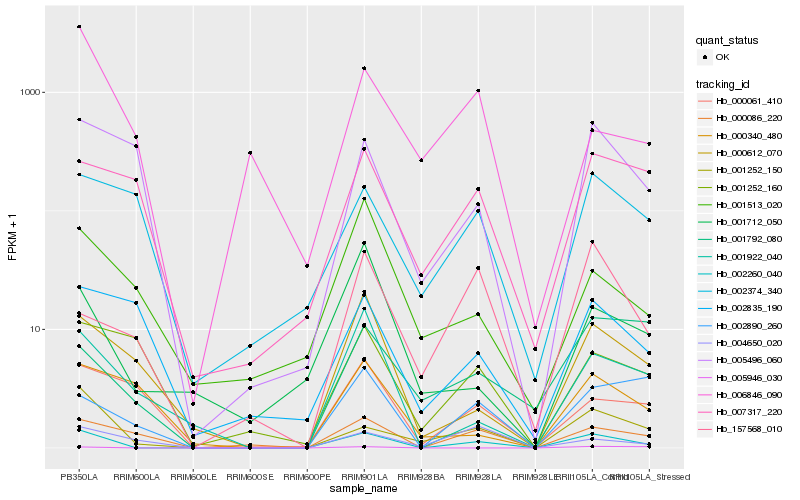
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002260_040 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 2 |
Hb_001252_150 |
0.2450497626 |
- |
- |
- |
| 3 |
Hb_000086_220 |
0.2777143608 |
- |
- |
- |
| 4 |
Hb_001922_040 |
0.2885100028 |
- |
- |
reticulon family protein [Populus trichocarpa] |
| 5 |
Hb_000612_070 |
0.293775671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002890_260 |
0.3104745866 |
- |
- |
- |
| 7 |
Hb_005496_060 |
0.3190684372 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 38-like [Jatropha curcas] |
| 8 |
Hb_000061_410 |
0.3288699748 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_005946_030 |
0.3326895684 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_001712_050 |
0.3328184646 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 11 |
Hb_157568_010 |
0.3328484029 |
- |
- |
MATE efflux family protein, putative [Theobroma cacao] |
| 12 |
Hb_007317_220 |
0.3354403343 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 13 |
Hb_004650_020 |
0.3357041184 |
- |
- |
- |
| 14 |
Hb_002374_340 |
0.3417474652 |
- |
- |
Two-component response regulator ARR9, putative [Ricinus communis] |
| 15 |
Hb_000340_480 |
0.3419586249 |
- |
- |
- |
| 16 |
Hb_001513_020 |
0.3432479931 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 17 |
Hb_001792_080 |
0.3448151472 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_001252_160 |
0.3458172958 |
- |
- |
PREDICTED: PRA1 family protein A3 [Jatropha curcas] |
| 19 |
Hb_002835_190 |
0.3468390297 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 20 |
Hb_006846_090 |
0.3471032845 |
- |
- |
PREDICTED: uncharacterized protein LOC105648695 [Jatropha curcas] |