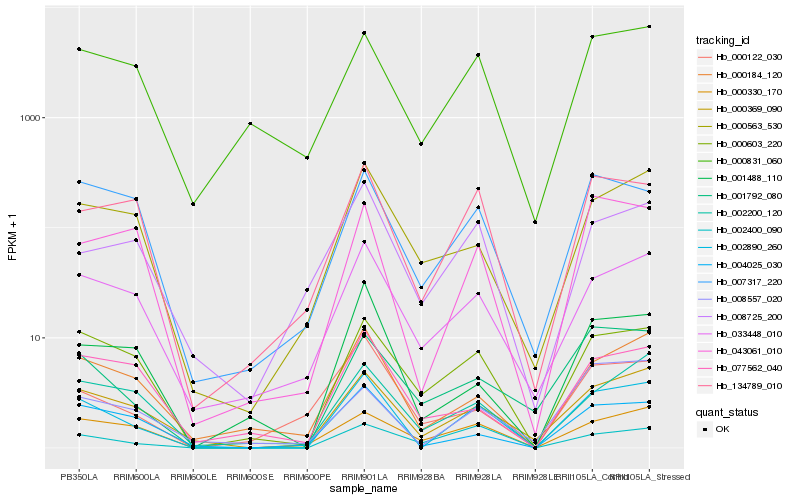
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_260 |
0.0 |
- |
- |
- |
| 2 |
Hb_000369_090 |
0.1701426116 |
- |
- |
- |
| 3 |
Hb_000603_220 |
0.1810783616 |
- |
- |
PREDICTED: MADS-box transcription factor 23 [Jatropha curcas] |
| 4 |
Hb_004025_030 |
0.1876790275 |
- |
- |
- |
| 5 |
Hb_000184_120 |
0.1892988209 |
- |
- |
hypothetical protein JCGZ_18526 [Jatropha curcas] |
| 6 |
Hb_043061_010 |
0.1895823252 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 7 |
Hb_033448_010 |
0.197737642 |
- |
- |
hypothetical protein JCGZ_19772 [Jatropha curcas] |
| 8 |
Hb_000330_170 |
0.1978566919 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 9 |
Hb_134789_010 |
0.1989468306 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_002400_090 |
0.2005968728 |
- |
- |
- |
| 11 |
Hb_001488_110 |
0.204281821 |
- |
- |
- |
| 12 |
Hb_002200_120 |
0.2053137103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001792_080 |
0.2056465194 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_000563_530 |
0.2117030867 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 15 |
Hb_007317_220 |
0.2143752758 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 16 |
Hb_008557_020 |
0.2212784116 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 17 |
Hb_000831_060 |
0.2251148985 |
- |
- |
17.5 kd heat shock family protein [Populus trichocarpa] |
| 18 |
Hb_008725_200 |
0.226811389 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 19 |
Hb_000122_030 |
0.2303840333 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 20 |
Hb_077562_040 |
0.2318771982 |
- |
- |
hypothetical protein JCGZ_24573 [Jatropha curcas] |