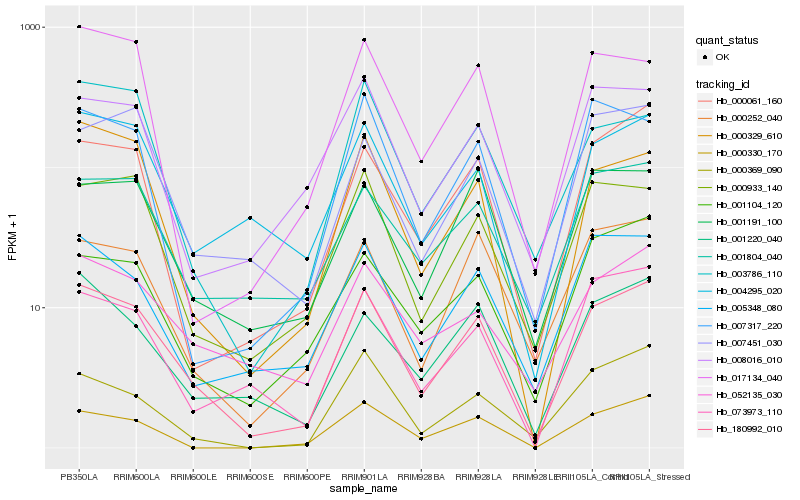
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180992_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s24060g [Populus trichocarpa] |
| 2 |
Hb_000329_610 |
0.1166065035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001220_040 |
0.128765846 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000933_140 |
0.1295030259 |
- |
- |
PREDICTED: uncharacterized protein LOC105630501 [Jatropha curcas] |
| 5 |
Hb_005348_080 |
0.1304167257 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |
| 6 |
Hb_004295_020 |
0.1305053244 |
- |
- |
thioredoxin II-type 1 [Hevea brasiliensis] |
| 7 |
Hb_000061_160 |
0.131220105 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000252_040 |
0.1330109945 |
- |
- |
PREDICTED: uncharacterized protein LOC105140231 [Populus euphratica] |
| 9 |
Hb_052135_030 |
0.1353547891 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 10 |
Hb_073973_110 |
0.1373242996 |
- |
- |
PREDICTED: uncharacterized protein LOC105638912 [Jatropha curcas] |
| 11 |
Hb_001191_100 |
0.1386303539 |
- |
- |
F-actin capping protein beta subunit [Populus trichocarpa] |
| 12 |
Hb_001104_120 |
0.1391817343 |
- |
- |
- |
| 13 |
Hb_001804_040 |
0.1398885706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003786_110 |
0.1439423699 |
- |
- |
PREDICTED: uncharacterized protein LOC105639617 [Jatropha curcas] |
| 15 |
Hb_007451_030 |
0.1449217668 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 16 |
Hb_000369_090 |
0.1455724307 |
- |
- |
- |
| 17 |
Hb_017134_040 |
0.1461420727 |
- |
- |
hypothetical protein AALP_AA3G042000 [Arabis alpina] |
| 18 |
Hb_007317_220 |
0.1475144193 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 19 |
Hb_000330_170 |
0.1479122398 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 20 |
Hb_008016_010 |
0.1482216293 |
- |
- |
PREDICTED: protein LURP-one-related 15-like isoform X1 [Jatropha curcas] |