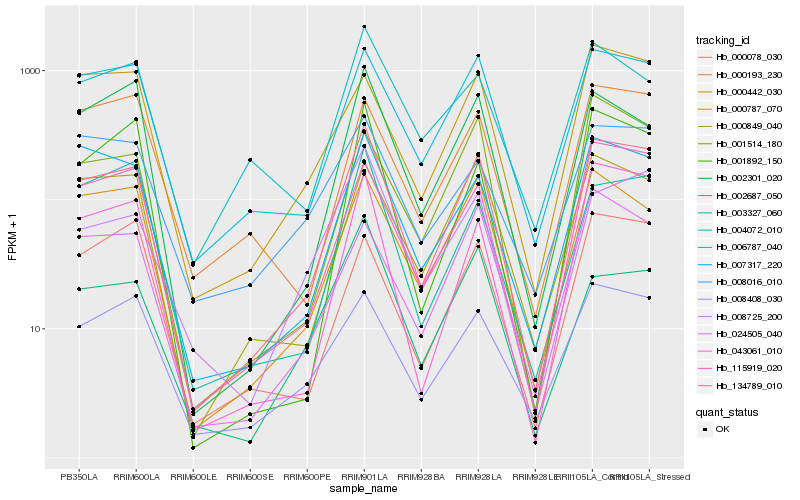
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134789_010 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_006787_040 |
0.1015070913 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 3 |
Hb_115919_020 |
0.1131440047 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_000442_030 |
0.1213876116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007317_220 |
0.1223810257 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 6 |
Hb_000849_040 |
0.1233070127 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_008408_030 |
0.1234113353 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 8 |
Hb_043061_010 |
0.1242885314 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 9 |
Hb_008725_200 |
0.1320393559 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 10 |
Hb_002301_020 |
0.1369954313 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 11 |
Hb_024505_040 |
0.1371814097 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |
| 12 |
Hb_001892_150 |
0.1381832968 |
- |
- |
- |
| 13 |
Hb_000787_070 |
0.1400138511 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 14 |
Hb_003327_060 |
0.1409031246 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000193_230 |
0.1422895891 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000078_030 |
0.1433022456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004072_010 |
0.144114638 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001514_180 |
0.14476484 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_16067 [Jatropha curcas] |
| 19 |
Hb_002687_050 |
0.1451584356 |
- |
- |
PREDICTED: uncharacterized protein LOC105782305 isoform X2 [Gossypium raimondii] |
| 20 |
Hb_008016_010 |
0.1451988346 |
- |
- |
PREDICTED: protein LURP-one-related 15-like isoform X1 [Jatropha curcas] |