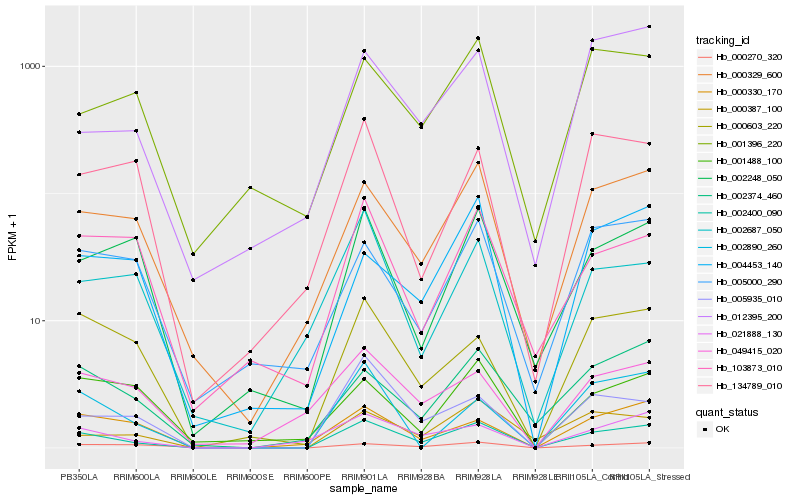
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000329_600 |
0.1634554926 |
- |
- |
PREDICTED: cell number regulator 6-like isoform X2 [Populus euphratica] |
| 3 |
Hb_021888_130 |
0.1771740892 |
- |
- |
- |
| 4 |
Hb_000603_220 |
0.1822011522 |
- |
- |
PREDICTED: MADS-box transcription factor 23 [Jatropha curcas] |
| 5 |
Hb_002374_460 |
0.1885841286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000330_170 |
0.1909310464 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 7 |
Hb_005935_010 |
0.1915733447 |
- |
- |
- |
| 8 |
Hb_002248_050 |
0.1974244692 |
- |
- |
PREDICTED: uncharacterized protein LOC105635697 [Jatropha curcas] |
| 9 |
Hb_000387_100 |
0.1994848368 |
- |
- |
- |
| 10 |
Hb_002890_260 |
0.2005968728 |
- |
- |
- |
| 11 |
Hb_000270_320 |
0.2041369572 |
- |
- |
hypothetical protein RCOM_1512920 [Ricinus communis] |
| 12 |
Hb_004453_140 |
0.2042666305 |
- |
- |
PREDICTED: uncharacterized protein LOC104880181 [Vitis vinifera] |
| 13 |
Hb_049415_020 |
0.2076827337 |
- |
- |
- |
| 14 |
Hb_103873_010 |
0.2102867447 |
- |
- |
hypothetical protein POPTR_0008s14260g [Populus trichocarpa] |
| 15 |
Hb_134789_010 |
0.210653794 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 16 |
Hb_005000_290 |
0.2123852383 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07430 [Jatropha curcas] |
| 17 |
Hb_012395_200 |
0.2132751571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002687_050 |
0.2158977679 |
- |
- |
PREDICTED: uncharacterized protein LOC105782305 isoform X2 [Gossypium raimondii] |
| 19 |
Hb_001396_220 |
0.2161927698 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 20 |
Hb_001488_100 |
0.2163823077 |
- |
- |
- |