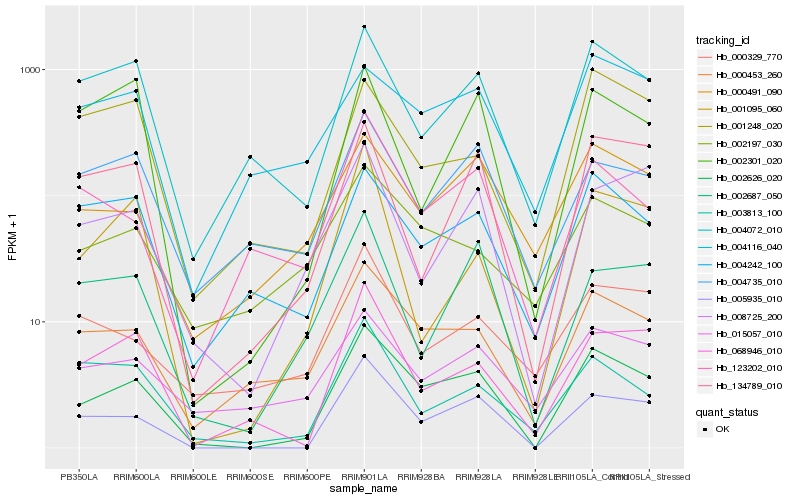
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002626_020 |
0.0 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 2 |
Hb_005935_010 |
0.1292853794 |
- |
- |
- |
| 3 |
Hb_000453_260 |
0.1634242421 |
- |
- |
PREDICTED: armadillo repeat-containing protein 7 isoform X1 [Vitis vinifera] |
| 4 |
Hb_004072_010 |
0.1870222509 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_068946_010 |
0.1888218317 |
- |
- |
hypothetical protein CICLE_v100122621mg, partial [Citrus clementina] |
| 6 |
Hb_000491_090 |
0.1955752485 |
- |
- |
hypothetical protein B456_008G219400 [Gossypium raimondii] |
| 7 |
Hb_134789_010 |
0.1958547463 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_004242_100 |
0.1986230942 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_008725_200 |
0.1990362512 |
- |
- |
PREDICTED: uncharacterized protein LOC105642590 [Jatropha curcas] |
| 10 |
Hb_123202_010 |
0.2008568037 |
- |
- |
thioredoxin h [Hevea brasiliensis] |
| 11 |
Hb_002687_050 |
0.2075922635 |
- |
- |
PREDICTED: uncharacterized protein LOC105782305 isoform X2 [Gossypium raimondii] |
| 12 |
Hb_015057_010 |
0.2080948304 |
- |
- |
PREDICTED: uncharacterized exonuclease domain-containing protein At3g15140 [Jatropha curcas] |
| 13 |
Hb_002197_030 |
0.2114499787 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_003813_100 |
0.2126656781 |
- |
- |
PREDICTED: proteasome subunit beta type-3-A [Cucumis sativus] |
| 15 |
Hb_004735_010 |
0.2130228729 |
- |
- |
PREDICTED: protein arginine N-methyltransferase PRMT10 [Vitis vinifera] |
| 16 |
Hb_001095_060 |
0.2131315081 |
- |
- |
Uncharacterized protein TCM_012505 [Theobroma cacao] |
| 17 |
Hb_000329_770 |
0.2168068424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004116_040 |
0.217927428 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0010s19720g [Populus trichocarpa] |
| 19 |
Hb_002301_020 |
0.2191246166 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 20 |
Hb_001248_020 |
0.219362267 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |