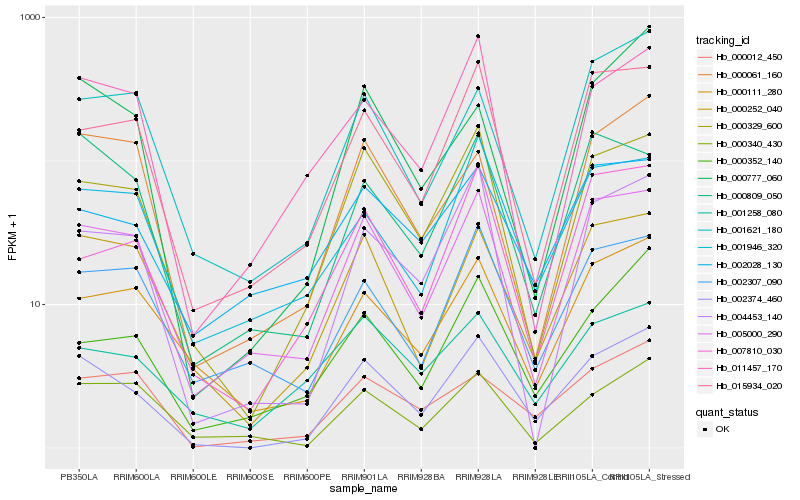
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_460 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000329_600 |
0.1222695228 |
- |
- |
PREDICTED: cell number regulator 6-like isoform X2 [Populus euphratica] |
| 3 |
Hb_005000_290 |
0.1334233669 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_07430 [Jatropha curcas] |
| 4 |
Hb_000252_040 |
0.1395364172 |
- |
- |
PREDICTED: uncharacterized protein LOC105140231 [Populus euphratica] |
| 5 |
Hb_015934_020 |
0.1505774661 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 6 |
Hb_004453_140 |
0.1574428825 |
- |
- |
PREDICTED: uncharacterized protein LOC104880181 [Vitis vinifera] |
| 7 |
Hb_000111_280 |
0.1591373085 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 8 |
Hb_000012_450 |
0.1599328562 |
- |
- |
PREDICTED: uncharacterized protein LOC105638155 [Jatropha curcas] |
| 9 |
Hb_000352_140 |
0.1607020604 |
- |
- |
- |
| 10 |
Hb_011457_170 |
0.1607766993 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 11 |
Hb_001946_320 |
0.1658150051 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 12 |
Hb_002307_090 |
0.1679834941 |
- |
- |
magnesium transporter CorA-like family protein [Populus trichocarpa] |
| 13 |
Hb_007810_030 |
0.1693926928 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 14 |
Hb_000777_060 |
0.1703507222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002028_130 |
0.1713226035 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 16 |
Hb_000061_160 |
0.1722974022 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000809_050 |
0.1726702549 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639663 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001621_180 |
0.1734659872 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-1 [Jatropha curcas] |
| 19 |
Hb_000340_430 |
0.1749050085 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001258_080 |
0.1752522741 |
- |
- |
PREDICTED: uncharacterized protein LOC105639921 [Jatropha curcas] |