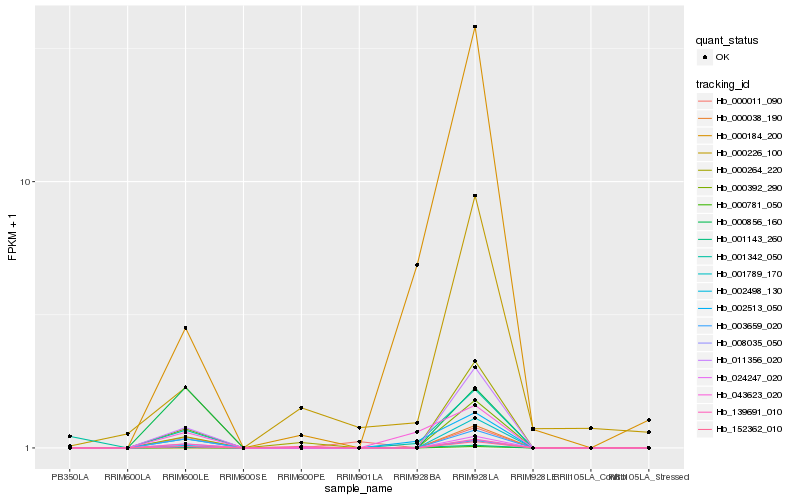
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 2 |
Hb_002498_130 |
0.0344146554 |
- |
- |
hypothetical protein POPTR_0004s00980g [Populus trichocarpa] |
| 3 |
Hb_000781_050 |
0.0413313737 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_000392_290 |
0.0435562335 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_011356_020 |
0.060137825 |
- |
- |
- |
| 6 |
Hb_139691_010 |
0.0930070364 |
- |
- |
- |
| 7 |
Hb_008035_050 |
0.1028313741 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 8 |
Hb_003659_020 |
0.1078921948 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 9 |
Hb_024247_020 |
0.1293292255 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 10 |
Hb_000264_220 |
0.2099350733 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |
| 11 |
Hb_000856_160 |
0.2568898384 |
- |
- |
- |
| 12 |
Hb_001143_260 |
0.2779651487 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 13 |
Hb_001342_050 |
0.2819831314 |
- |
- |
- |
| 14 |
Hb_152362_010 |
0.2998509375 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 15 |
Hb_000011_090 |
0.3131870707 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 16 |
Hb_000184_200 |
0.313679649 |
- |
- |
- |
| 17 |
Hb_000226_100 |
0.3224142241 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_043623_020 |
0.3306025526 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 19 |
Hb_002513_050 |
0.3330004502 |
- |
- |
hypothetical protein F383_14794 [Gossypium arboreum] |
| 20 |
Hb_000038_190 |
0.3425373194 |
- |
- |
- |