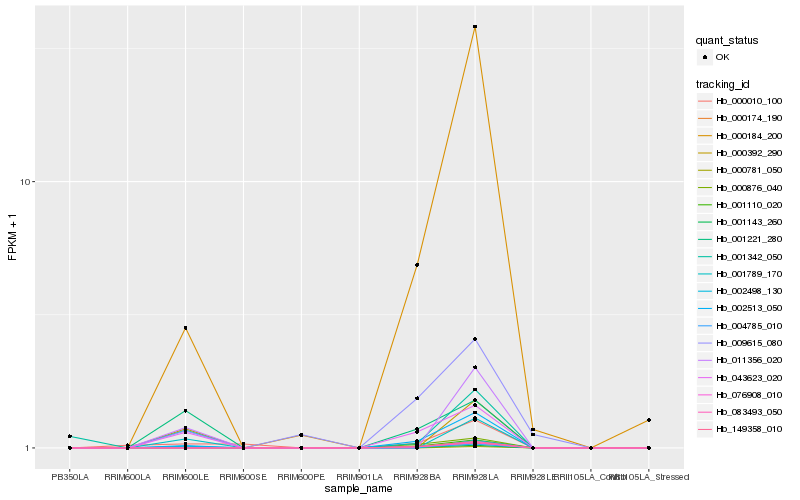
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_260 |
0.0 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 2 |
Hb_043623_020 |
0.0839114657 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 3 |
Hb_002513_050 |
0.1641632416 |
- |
- |
hypothetical protein F383_14794 [Gossypium arboreum] |
| 4 |
Hb_000876_040 |
0.1703713439 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 5 |
Hb_001221_280 |
0.2072438986 |
- |
- |
- |
| 6 |
Hb_000184_200 |
0.208492365 |
- |
- |
- |
| 7 |
Hb_009615_080 |
0.2656321485 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 8 |
Hb_002498_130 |
0.2763255755 |
- |
- |
hypothetical protein POPTR_0004s00980g [Populus trichocarpa] |
| 9 |
Hb_000781_050 |
0.2764719055 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_001342_050 |
0.2770748478 |
- |
- |
- |
| 11 |
Hb_011356_020 |
0.2776785239 |
- |
- |
- |
| 12 |
Hb_001789_170 |
0.2779651487 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 13 |
Hb_000010_100 |
0.2853343265 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 14 |
Hb_000392_290 |
0.2855241711 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_004785_010 |
0.2907578068 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_000174_190 |
0.2907714677 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_076908_010 |
0.2907793863 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 18 |
Hb_083493_050 |
0.2907861216 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 19 |
Hb_149358_010 |
0.2908080681 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 20 |
Hb_001110_020 |
0.2908566017 |
- |
- |
hypothetical protein JCGZ_03693 [Jatropha curcas] |