| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
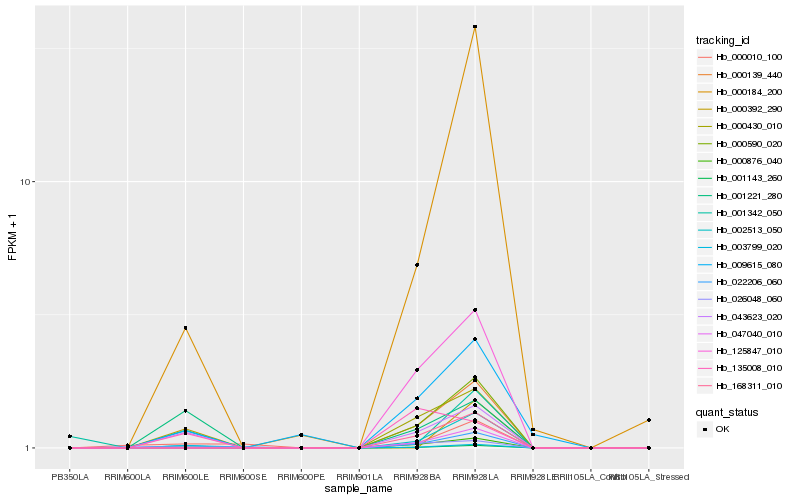
Hb_043623_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 2 |
Hb_001143_260 |
0.0839114657 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 3 |
Hb_001221_280 |
0.1534331645 |
- |
- |
- |
| 4 |
Hb_000876_040 |
0.1871371915 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 5 |
Hb_002513_050 |
0.2268280112 |
- |
- |
hypothetical protein F383_14794 [Gossypium arboreum] |
| 6 |
Hb_009615_080 |
0.2733063028 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 7 |
Hb_000184_200 |
0.2775530385 |
- |
- |
- |
| 8 |
Hb_135008_010 |
0.2902501573 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 9 |
Hb_003799_020 |
0.2935763526 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 10 |
Hb_001342_050 |
0.3133018813 |
- |
- |
- |
| 11 |
Hb_000010_100 |
0.3155173887 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 12 |
Hb_168311_010 |
0.324781984 |
- |
- |
- |
| 13 |
Hb_000139_440 |
0.326347058 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_125847_010 |
0.3271167476 |
- |
- |
- |
| 15 |
Hb_000590_020 |
0.3275860177 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 16 |
Hb_047040_010 |
0.3282137223 |
- |
- |
integrase [Gossypium herbaceum] |
| 17 |
Hb_026048_060 |
0.328902539 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 18 |
Hb_000430_010 |
0.3293738443 |
- |
- |
- |
| 19 |
Hb_022206_060 |
0.3294783301 |
- |
- |
- |
| 20 |
Hb_000392_290 |
0.3297629429 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |