| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
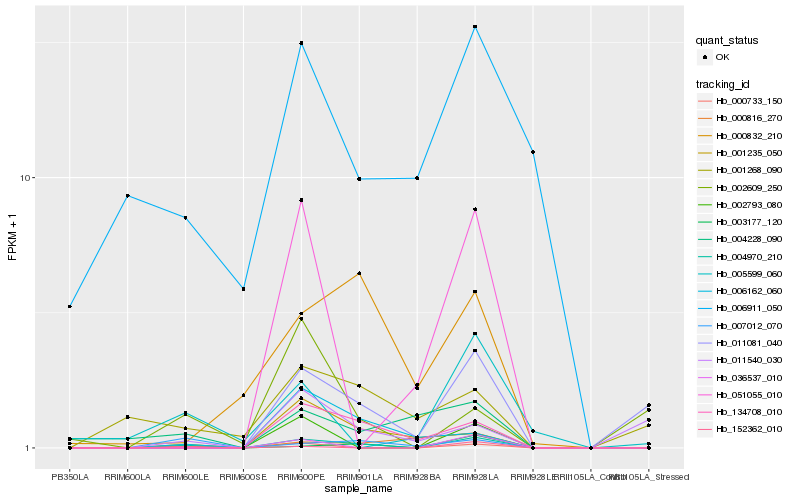
Hb_004970_210 |
0.0 |
- |
- |
- |
| 2 |
Hb_134708_010 |
0.3342963344 |
- |
- |
PREDICTED: uncharacterized protein LOC102604542 [Solanum tuberosum] |
| 3 |
Hb_007012_070 |
0.3433288859 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 4 |
Hb_152362_010 |
0.3543193211 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 5 |
Hb_001235_050 |
0.3630315861 |
- |
- |
PREDICTED: proteasome activator subunit 4 [Nelumbo nucifera] |
| 6 |
Hb_011081_040 |
0.3638225486 |
- |
- |
hypothetical protein JCGZ_20956 [Jatropha curcas] |
| 7 |
Hb_003177_120 |
0.3729576368 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Populus euphratica] |
| 8 |
Hb_036537_010 |
0.3761557933 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_006162_060 |
0.3827839287 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 10 |
Hb_000733_150 |
0.383115243 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Populus euphratica] |
| 11 |
Hb_002793_080 |
0.38522417 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 12 |
Hb_000832_210 |
0.3923447185 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 13 |
Hb_051055_010 |
0.4024910052 |
- |
- |
- |
| 14 |
Hb_005599_060 |
0.4042966037 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002609_250 |
0.4055582066 |
- |
- |
PREDICTED: starch-binding domain-containing protein 1-like [Jatropha curcas] |
| 16 |
Hb_000816_270 |
0.4069242187 |
- |
- |
- |
| 17 |
Hb_006911_050 |
0.4093952072 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 18 |
Hb_004228_090 |
0.4098370795 |
- |
- |
- |
| 19 |
Hb_001268_090 |
0.4132618426 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 20 |
Hb_011540_030 |
0.416341473 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |