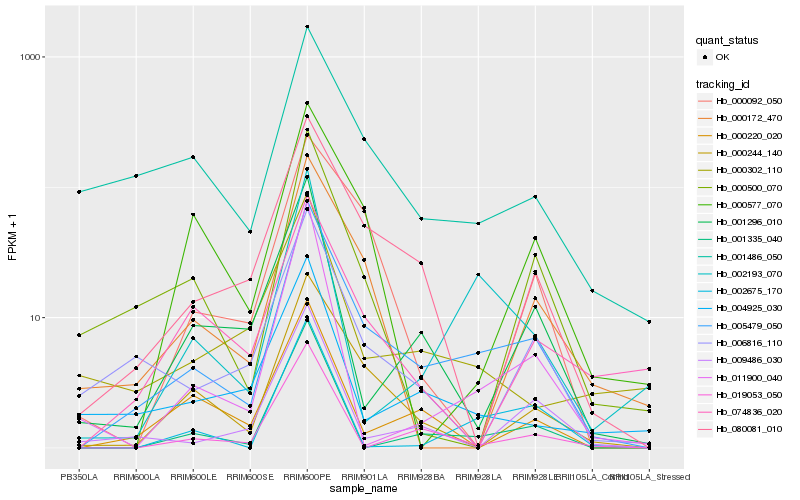
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005479_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_080081_010 |
0.181252758 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_000244_140 |
0.1872537154 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |
| 4 |
Hb_001486_050 |
0.2080572422 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 5 |
Hb_006816_110 |
0.2156321047 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 6 |
Hb_000172_470 |
0.2184202798 |
- |
- |
hypothetical protein POPTR_0005s08310g [Populus trichocarpa] |
| 7 |
Hb_001335_040 |
0.2202832245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000092_050 |
0.22839836 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 9 |
Hb_000577_070 |
0.2293821881 |
- |
- |
- |
| 10 |
Hb_000500_070 |
0.2309977396 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 11 |
Hb_001296_010 |
0.2328283586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000220_020 |
0.2337426629 |
- |
- |
hypothetical protein CICLE_v10017575mg [Citrus clementina] |
| 13 |
Hb_002193_070 |
0.2345484777 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_009486_030 |
0.2362733583 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Jatropha curcas] |
| 15 |
Hb_002675_170 |
0.2376527328 |
- |
- |
hypothetical protein JCGZ_06235 [Jatropha curcas] |
| 16 |
Hb_004925_030 |
0.2386985238 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 17 |
Hb_011900_040 |
0.2396624706 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 18 |
Hb_074836_020 |
0.2405787289 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000302_110 |
0.2417937058 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_019053_050 |
0.2429156354 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |