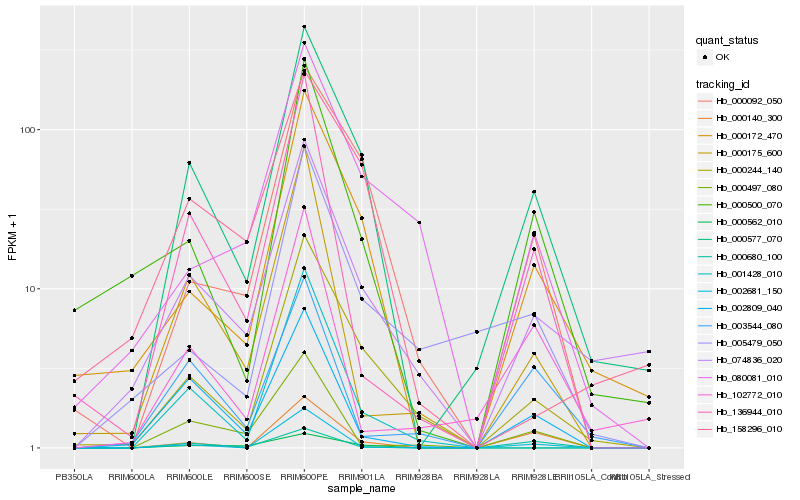
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000577_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000244_140 |
0.145406172 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |
| 3 |
Hb_000172_470 |
0.1469942381 |
- |
- |
hypothetical protein POPTR_0005s08310g [Populus trichocarpa] |
| 4 |
Hb_000092_050 |
0.1760693743 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 5 |
Hb_074836_020 |
0.1767849394 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000140_300 |
0.2008618347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000500_070 |
0.2045265301 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 8 |
Hb_002809_040 |
0.212466147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_158296_010 |
0.2244919065 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 10 |
Hb_136944_010 |
0.2250646166 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 11 |
Hb_002681_150 |
0.226070207 |
- |
- |
uridine cytidine kinase I, putative [Ricinus communis] |
| 12 |
Hb_005479_050 |
0.2293821881 |
- |
- |
- |
| 13 |
Hb_080081_010 |
0.2337937358 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Fragaria vesca subsp. vesca] |
| 14 |
Hb_001428_010 |
0.2363987801 |
- |
- |
PREDICTED: transcription factor PRE1-like [Jatropha curcas] |
| 15 |
Hb_102772_010 |
0.2366121086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000680_100 |
0.2371794953 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_000562_010 |
0.2413738254 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 18 |
Hb_000175_600 |
0.2440881515 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 19 |
Hb_000497_080 |
0.2450096008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003544_080 |
0.2486457182 |
- |
- |
Protein HVA22, putative [Ricinus communis] |