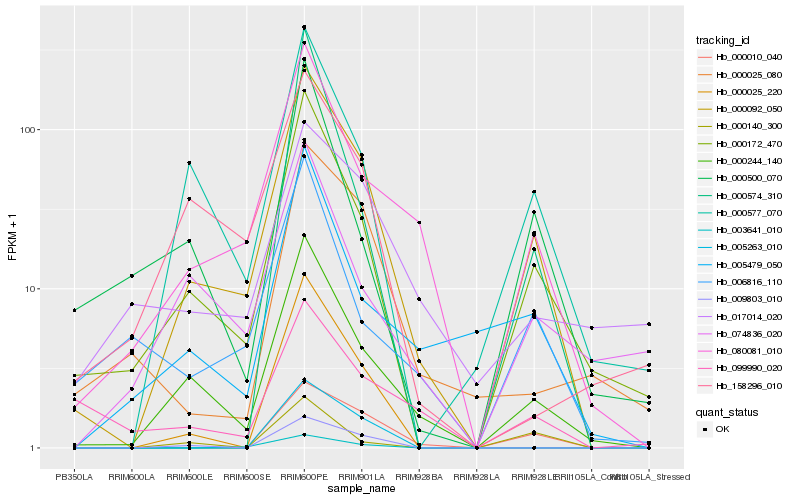
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000092_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 2 |
Hb_000244_140 |
0.139607031 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |
| 3 |
Hb_000172_470 |
0.1508856201 |
- |
- |
hypothetical protein POPTR_0005s08310g [Populus trichocarpa] |
| 4 |
Hb_080081_010 |
0.1674520017 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Fragaria vesca subsp. vesca] |
| 5 |
Hb_000577_070 |
0.1760693743 |
- |
- |
- |
| 6 |
Hb_000010_040 |
0.19767284 |
- |
- |
PREDICTED: probable 1-aminocyclopropane-1-carboxylate deaminase-like isoform X2 [Citrus sinensis] |
| 7 |
Hb_000025_220 |
0.2221778471 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 8 |
Hb_009803_010 |
0.2242539398 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 9 |
Hb_005479_050 |
0.22839836 |
- |
- |
- |
| 10 |
Hb_003641_010 |
0.2329280228 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 11 |
Hb_006816_110 |
0.2387063973 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 12 |
Hb_158296_010 |
0.2388596331 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 13 |
Hb_074836_020 |
0.2438562438 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000140_300 |
0.2449265871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000500_070 |
0.2458242439 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 16 |
Hb_000025_080 |
0.2465827823 |
- |
- |
- |
| 17 |
Hb_000574_310 |
0.2498814954 |
- |
- |
hypothetical protein CISIN_1g027539mg [Citrus sinensis] |
| 18 |
Hb_005263_010 |
0.2517452052 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 19 |
Hb_017014_020 |
0.2608566643 |
- |
- |
- |
| 20 |
Hb_099990_020 |
0.2634568371 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |