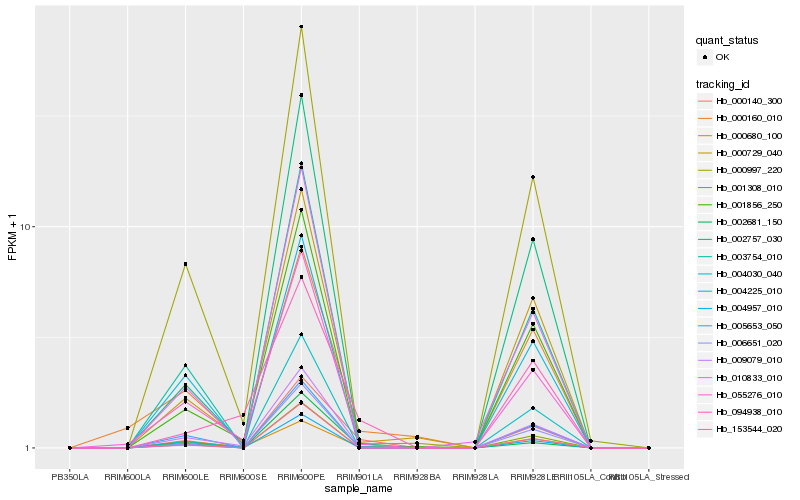
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_300 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009079_010 |
0.1603662388 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 3 |
Hb_055276_010 |
0.1721864907 |
- |
- |
hypothetical protein POPTR_0003s02950g [Populus trichocarpa] |
| 4 |
Hb_000729_040 |
0.1738223332 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 5 |
Hb_003754_010 |
0.1747537746 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 6 |
Hb_002681_150 |
0.1764553345 |
- |
- |
uridine cytidine kinase I, putative [Ricinus communis] |
| 7 |
Hb_000997_220 |
0.1783041581 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 8 |
Hb_004957_010 |
0.1790540299 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 9 |
Hb_001308_010 |
0.1794491157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006651_020 |
0.1807003756 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 11 |
Hb_000160_010 |
0.1823614909 |
- |
- |
PREDICTED: uncharacterized protein LOC105638816 [Jatropha curcas] |
| 12 |
Hb_153544_020 |
0.1833002026 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 13 |
Hb_000680_100 |
0.1838838286 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_002757_030 |
0.1865868699 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_094938_010 |
0.1874508372 |
- |
- |
- |
| 16 |
Hb_010833_010 |
0.1877440379 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 17 |
Hb_004030_040 |
0.188504941 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 18 |
Hb_005653_050 |
0.1893220214 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 19 |
Hb_004225_010 |
0.1897123718 |
- |
- |
- |
| 20 |
Hb_001856_250 |
0.1898081301 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |