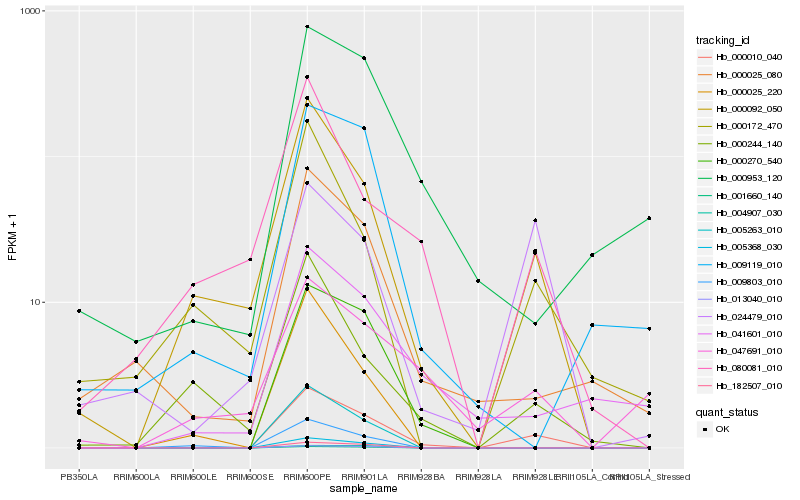
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_040 |
0.0 |
- |
- |
PREDICTED: probable 1-aminocyclopropane-1-carboxylate deaminase-like isoform X2 [Citrus sinensis] |
| 2 |
Hb_000092_050 |
0.19767284 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 3 |
Hb_000270_540 |
0.2259517478 |
- |
- |
hypothetical protein JCGZ_11527 [Jatropha curcas] |
| 4 |
Hb_005368_030 |
0.2382840034 |
- |
- |
- |
| 5 |
Hb_005263_010 |
0.2432623077 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 6 |
Hb_182507_010 |
0.2484403741 |
- |
- |
- |
| 7 |
Hb_001660_140 |
0.2494010088 |
- |
- |
- |
| 8 |
Hb_000025_080 |
0.2495304497 |
- |
- |
- |
| 9 |
Hb_013040_010 |
0.2565827671 |
- |
- |
hypothetical protein POPTR_0009s09690g [Populus trichocarpa] |
| 10 |
Hb_024479_010 |
0.2576108338 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 11 |
Hb_041601_010 |
0.2600447132 |
- |
- |
- |
| 12 |
Hb_000953_120 |
0.2734467614 |
- |
- |
- |
| 13 |
Hb_000244_140 |
0.2748425431 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |
| 14 |
Hb_080081_010 |
0.2783730125 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Fragaria vesca subsp. vesca] |
| 15 |
Hb_009803_010 |
0.2803578874 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 16 |
Hb_000025_220 |
0.2831836519 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 17 |
Hb_004907_030 |
0.2832033265 |
- |
- |
hypothetical protein POPTR_0012s032802g, partial [Populus trichocarpa] |
| 18 |
Hb_009119_010 |
0.2837170527 |
- |
- |
- |
| 19 |
Hb_000172_470 |
0.2839458829 |
- |
- |
hypothetical protein POPTR_0005s08310g [Populus trichocarpa] |
| 20 |
Hb_047691_010 |
0.2890148147 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |