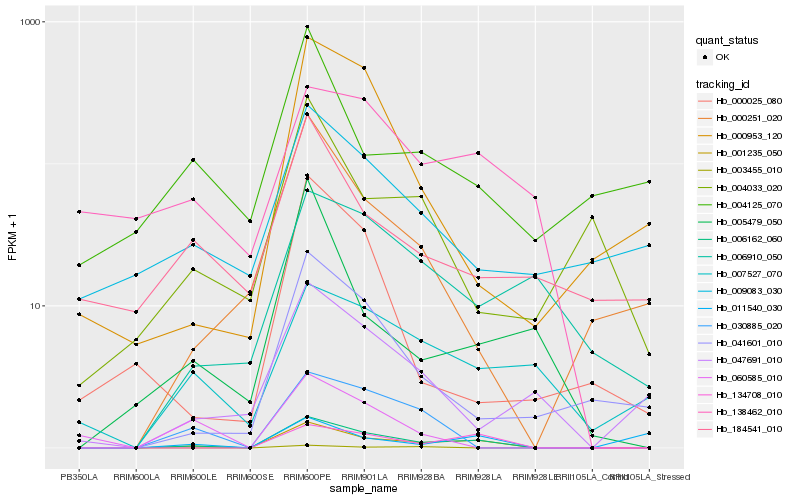
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_050 |
0.0 |
- |
- |
PREDICTED: proteasome activator subunit 4 [Nelumbo nucifera] |
| 2 |
Hb_006162_060 |
0.0646432906 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 3 |
Hb_134708_010 |
0.1910620475 |
- |
- |
PREDICTED: uncharacterized protein LOC102604542 [Solanum tuberosum] |
| 4 |
Hb_030885_020 |
0.3064973239 |
- |
- |
- |
| 5 |
Hb_041601_010 |
0.3065675833 |
- |
- |
- |
| 6 |
Hb_000251_020 |
0.3079225351 |
- |
- |
hypothetical protein CISIN_1g028541mg [Citrus sinensis] |
| 7 |
Hb_005479_050 |
0.3180408828 |
- |
- |
- |
| 8 |
Hb_007527_070 |
0.3201525293 |
- |
- |
PREDICTED: uncharacterized protein LOC105637197 [Jatropha curcas] |
| 9 |
Hb_006910_050 |
0.3224889289 |
- |
- |
hypothetical protein CISIN_1g0480481mg, partial [Citrus sinensis] |
| 10 |
Hb_138462_010 |
0.3241892911 |
- |
- |
PREDICTED: SUMO-conjugating enzyme UBC9-A-like isoform X1 [Elaeis guineensis] |
| 11 |
Hb_000953_120 |
0.3273988896 |
- |
- |
- |
| 12 |
Hb_011540_030 |
0.3335652542 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX5-like isoform X6 [Glycine max] |
| 13 |
Hb_047691_010 |
0.339191097 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 14 |
Hb_004033_020 |
0.3395408518 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 15 |
Hb_003455_010 |
0.3398856532 |
- |
- |
hypothetical protein POPTR_0009s14040g [Populus trichocarpa] |
| 16 |
Hb_184541_010 |
0.3452480434 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 17 |
Hb_004125_070 |
0.3484485446 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 18 |
Hb_009083_030 |
0.3491258754 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 19 |
Hb_000025_080 |
0.3494633232 |
- |
- |
- |
| 20 |
Hb_060585_010 |
0.3505590481 |
- |
- |
unnamed protein product [Coffea canephora] |