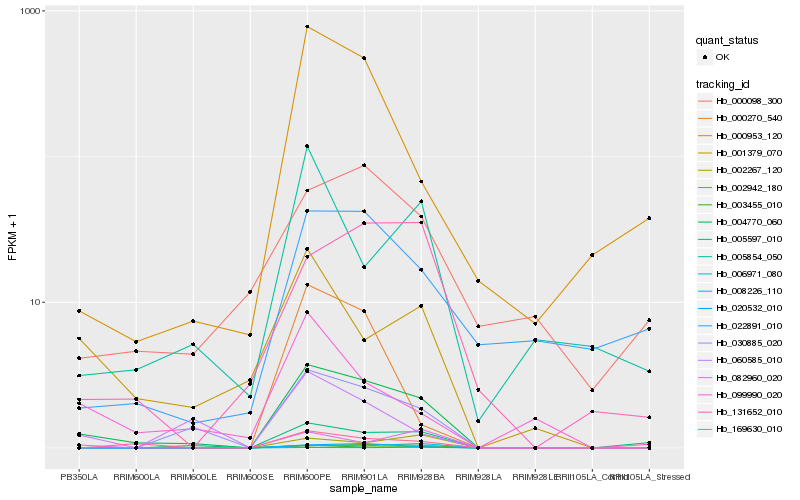
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004770_060 |
0.0 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 2 |
Hb_030885_020 |
0.2139324791 |
- |
- |
- |
| 3 |
Hb_020532_010 |
0.2389571565 |
- |
- |
PREDICTED: uncharacterized protein LOC105775165 [Gossypium raimondii] |
| 4 |
Hb_169630_010 |
0.242306762 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 5 |
Hb_003455_010 |
0.243163144 |
- |
- |
hypothetical protein POPTR_0009s14040g [Populus trichocarpa] |
| 6 |
Hb_002942_180 |
0.2638370686 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_005597_010 |
0.2726286255 |
- |
- |
- |
| 8 |
Hb_001379_070 |
0.2824401244 |
- |
- |
hypothetical protein JCGZ_16377 [Jatropha curcas] |
| 9 |
Hb_000953_120 |
0.2857560497 |
- |
- |
- |
| 10 |
Hb_060585_010 |
0.2875736296 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_131652_010 |
0.2951219982 |
- |
- |
hypothetical protein CICLE_v10010066mg [Citrus clementina] |
| 12 |
Hb_082960_020 |
0.2978042522 |
- |
- |
hypothetical protein B456_012G107300 [Gossypium raimondii] |
| 13 |
Hb_006971_080 |
0.3020614294 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 14 |
Hb_002267_120 |
0.305636845 |
- |
- |
hypothetical protein B456_009G136300 [Gossypium raimondii] |
| 15 |
Hb_000098_300 |
0.3107692532 |
- |
- |
- |
| 16 |
Hb_022891_010 |
0.3117009849 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 17 |
Hb_005854_050 |
0.3123027496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_099990_020 |
0.3142388156 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 19 |
Hb_000270_540 |
0.3159583974 |
- |
- |
hypothetical protein JCGZ_11527 [Jatropha curcas] |
| 20 |
Hb_008226_110 |
0.3167513219 |
- |
- |
uncharacterized protein LOC100501343 [Zea mays] |