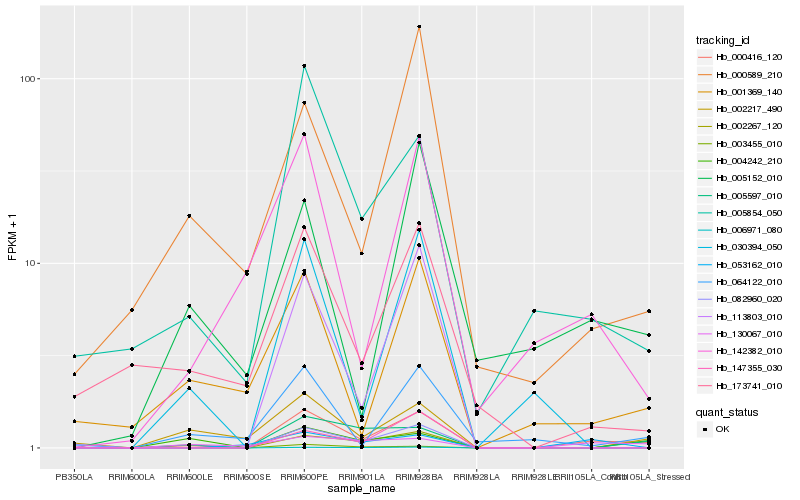
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000416_120 |
0.0 |
- |
- |
Putative LRR receptor-like serine/threonine-protein kinase, partial [Glycine soja] |
| 2 |
Hb_005854_050 |
0.2571756333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005597_010 |
0.2782795556 |
- |
- |
- |
| 4 |
Hb_142382_010 |
0.2805975326 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |
| 5 |
Hb_113803_010 |
0.2885474054 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002267_120 |
0.2933165114 |
- |
- |
hypothetical protein B456_009G136300 [Gossypium raimondii] |
| 7 |
Hb_130067_010 |
0.2942344328 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_001369_140 |
0.2978465857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_064122_010 |
0.2985004056 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005152_010 |
0.3019488955 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 11 |
Hb_006971_080 |
0.3021936389 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_030394_050 |
0.3043404164 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_173741_010 |
0.3065130005 |
- |
- |
PREDICTED: caffeoyl-CoA O-methyltransferase isoform X3 [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_000589_210 |
0.3071148058 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 15 |
Hb_003455_010 |
0.3071751057 |
- |
- |
hypothetical protein POPTR_0009s14040g [Populus trichocarpa] |
| 16 |
Hb_002217_490 |
0.3075335526 |
- |
- |
- |
| 17 |
Hb_147355_030 |
0.3134391987 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 isoform X5 [Populus euphratica] |
| 18 |
Hb_082960_020 |
0.3152325763 |
- |
- |
hypothetical protein B456_012G107300 [Gossypium raimondii] |
| 19 |
Hb_053162_010 |
0.3152564134 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 20 |
Hb_004242_210 |
0.316170258 |
- |
- |
PREDICTED: uncharacterized protein LOC105633248 [Jatropha curcas] |