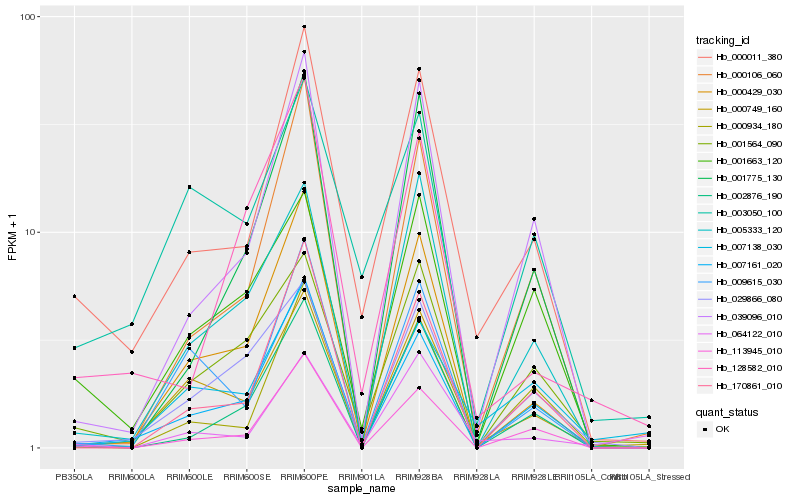
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_380 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 2 |
Hb_007161_020 |
0.1566453905 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 3 |
Hb_039096_010 |
0.1622706523 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_113945_010 |
0.1680492217 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000749_160 |
0.1709554271 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 6 |
Hb_000429_030 |
0.1747181282 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000106_060 |
0.1748542152 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 8 |
Hb_001564_090 |
0.1826347046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_170861_010 |
0.1842837555 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003050_100 |
0.1865807853 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 11 |
Hb_001663_120 |
0.1914910049 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002876_190 |
0.1920384607 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 13 |
Hb_128582_010 |
0.1926509498 |
- |
- |
- |
| 14 |
Hb_009615_030 |
0.1926946018 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_005333_120 |
0.1934178295 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 16 |
Hb_007138_030 |
0.1941555077 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 17 |
Hb_000934_180 |
0.1950623738 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 18 |
Hb_001775_130 |
0.1962167 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 19 |
Hb_064122_010 |
0.1964152525 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 20 |
Hb_029866_080 |
0.1972084049 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |