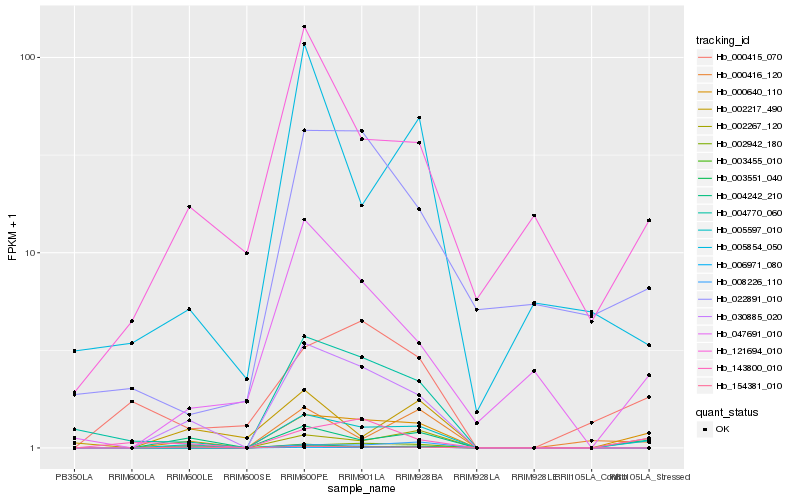
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005597_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000640_110 |
0.1106353826 |
- |
- |
- |
| 3 |
Hb_004242_210 |
0.222110183 |
- |
- |
PREDICTED: uncharacterized protein LOC105633248 [Jatropha curcas] |
| 4 |
Hb_030885_020 |
0.2311959439 |
- |
- |
- |
| 5 |
Hb_003455_010 |
0.2618961617 |
- |
- |
hypothetical protein POPTR_0009s14040g [Populus trichocarpa] |
| 6 |
Hb_004770_060 |
0.2726286255 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 7 |
Hb_000416_120 |
0.2782795556 |
- |
- |
Putative LRR receptor-like serine/threonine-protein kinase, partial [Glycine soja] |
| 8 |
Hb_047691_010 |
0.2881801325 |
- |
- |
Uncharacterized protein F383_01974 [Gossypium arboreum] |
| 9 |
Hb_006971_080 |
0.2912616578 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_002267_120 |
0.2922924733 |
- |
- |
hypothetical protein B456_009G136300 [Gossypium raimondii] |
| 11 |
Hb_154381_010 |
0.2942055605 |
- |
- |
- |
| 12 |
Hb_003551_040 |
0.2990804683 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 13 |
Hb_002942_180 |
0.3028460391 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 14 |
Hb_143800_010 |
0.3059569356 |
- |
- |
hypothetical protein POPTR_0010s21410g, partial [Populus trichocarpa] |
| 15 |
Hb_005854_050 |
0.307084709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008226_110 |
0.3093220994 |
- |
- |
uncharacterized protein LOC100501343 [Zea mays] |
| 17 |
Hb_121694_010 |
0.3108795574 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 18 |
Hb_022891_010 |
0.3111487882 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 19 |
Hb_002217_490 |
0.3132047086 |
- |
- |
- |
| 20 |
Hb_000415_070 |
0.3148964279 |
- |
- |
- |