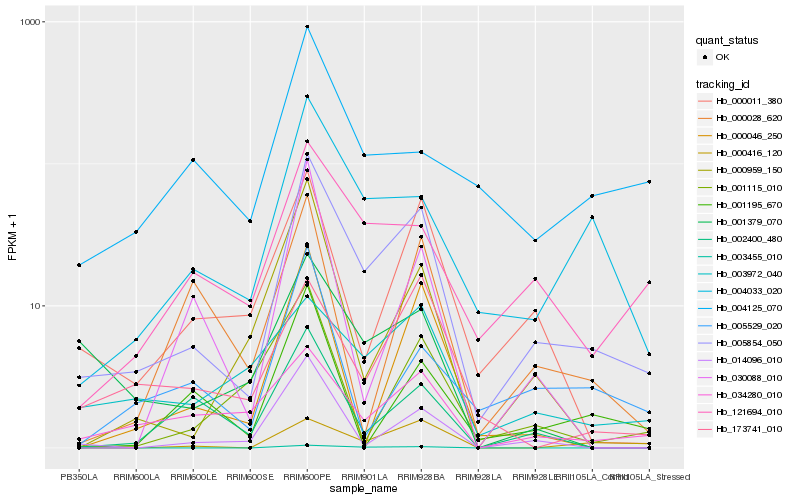
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005854_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004033_020 |
0.2062941021 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 3 |
Hb_121694_010 |
0.2262437679 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 4 |
Hb_000011_380 |
0.2362780951 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 5 |
Hb_034280_010 |
0.2426078137 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 6 |
Hb_014096_010 |
0.2490825161 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_003455_010 |
0.2513913305 |
- |
- |
hypothetical protein POPTR_0009s14040g [Populus trichocarpa] |
| 8 |
Hb_000416_120 |
0.2571756333 |
- |
- |
Putative LRR receptor-like serine/threonine-protein kinase, partial [Glycine soja] |
| 9 |
Hb_003972_040 |
0.2591096574 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 10 |
Hb_000959_150 |
0.2601531543 |
- |
- |
PREDICTED: uncharacterized protein LOC104879927 [Vitis vinifera] |
| 11 |
Hb_002400_480 |
0.2612484599 |
- |
- |
PREDICTED: uncharacterized protein LOC105637596 [Jatropha curcas] |
| 12 |
Hb_001379_070 |
0.2635655568 |
- |
- |
hypothetical protein JCGZ_16377 [Jatropha curcas] |
| 13 |
Hb_173741_010 |
0.2638311945 |
- |
- |
PREDICTED: caffeoyl-CoA O-methyltransferase isoform X3 [Musa acuminata subsp. malaccensis] |
| 14 |
Hb_000046_250 |
0.2676512966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005529_020 |
0.2688034991 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 16 |
Hb_004125_070 |
0.2688357768 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 17 |
Hb_001195_670 |
0.2717627269 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 18 |
Hb_001115_010 |
0.2734097864 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 2-like [Jatropha curcas] |
| 19 |
Hb_030088_010 |
0.2762556791 |
- |
- |
basic blue copper family protein [Populus trichocarpa] |
| 20 |
Hb_000028_620 |
0.2776017892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |