| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
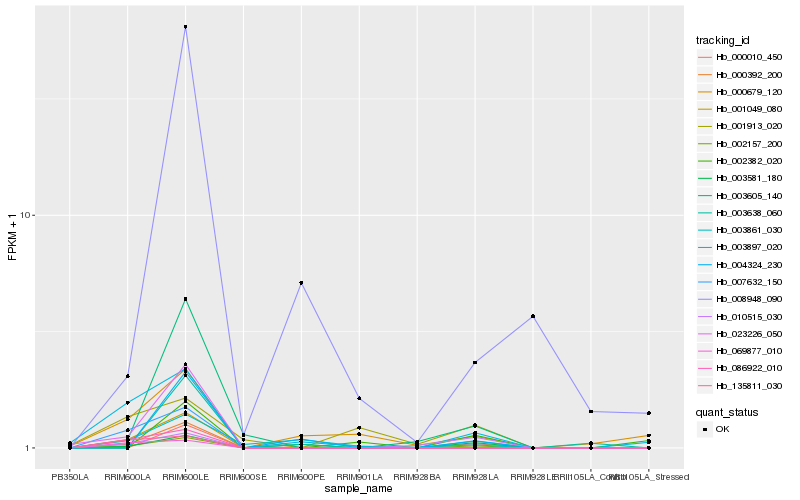
Hb_002157_200 |
0.0 |
- |
- |
hypothetical protein VITISV_040170 [Vitis vinifera] |
| 2 |
Hb_003897_020 |
0.2787260744 |
transcription factor |
TF Family: Orphans |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_003605_140 |
0.2962935873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001049_080 |
0.297881887 |
- |
- |
- |
| 5 |
Hb_000392_200 |
0.3004647399 |
- |
- |
PREDICTED: cytochrome P450 98A2-like isoform X2 [Phoenix dactylifera] |
| 6 |
Hb_000010_450 |
0.3125420243 |
- |
- |
purine permease, putative [Ricinus communis] |
| 7 |
Hb_010515_030 |
0.318614858 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 8 |
Hb_000679_120 |
0.3228523624 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 9 |
Hb_007632_150 |
0.323412778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_023226_050 |
0.3261199707 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 11 |
Hb_086922_010 |
0.3301396254 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 12 |
Hb_002382_020 |
0.3353921826 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 13 |
Hb_001913_020 |
0.3370532308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003861_030 |
0.3393190315 |
- |
- |
hypothetical protein JCGZ_16321 [Jatropha curcas] |
| 15 |
Hb_003581_180 |
0.3396688324 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 16 |
Hb_003638_060 |
0.3617551367 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 17 |
Hb_004324_230 |
0.3634023044 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 18 |
Hb_008948_090 |
0.3680646798 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_135811_030 |
0.3700415359 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_069877_010 |
0.372617956 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |