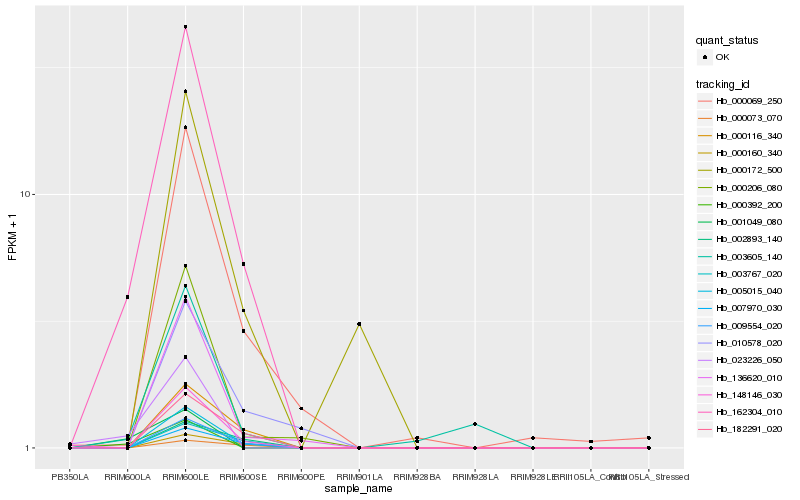
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162304_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005015_040 |
0.170962836 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1-like [Nicotiana sylvestris] |
| 3 |
Hb_009554_020 |
0.174893039 |
- |
- |
PREDICTED: em-like protein GEA6 [Sesamum indicum] |
| 4 |
Hb_007970_030 |
0.2047084085 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_148146_030 |
0.2061230035 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 6 |
Hb_182291_020 |
0.206356438 |
- |
- |
hypothetical protein CICLE_v10029871mg [Citrus clementina] |
| 7 |
Hb_000116_340 |
0.2082841217 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 8 |
Hb_000069_250 |
0.2111668576 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 9 |
Hb_000392_200 |
0.219595613 |
- |
- |
PREDICTED: cytochrome P450 98A2-like isoform X2 [Phoenix dactylifera] |
| 10 |
Hb_136620_010 |
0.2207177115 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 11 |
Hb_003767_020 |
0.2227546362 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_003605_140 |
0.2284099285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000160_340 |
0.228454289 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000206_080 |
0.2312805167 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 15 |
Hb_023226_050 |
0.2359082562 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 16 |
Hb_002893_140 |
0.2389004713 |
- |
- |
Glycine-rich cell wall structural protein precursor, putative [Ricinus communis] |
| 17 |
Hb_010578_020 |
0.243986712 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001049_080 |
0.2518346965 |
- |
- |
- |
| 19 |
Hb_000172_500 |
0.2530427235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000073_070 |
0.2568485677 |
- |
- |
- |