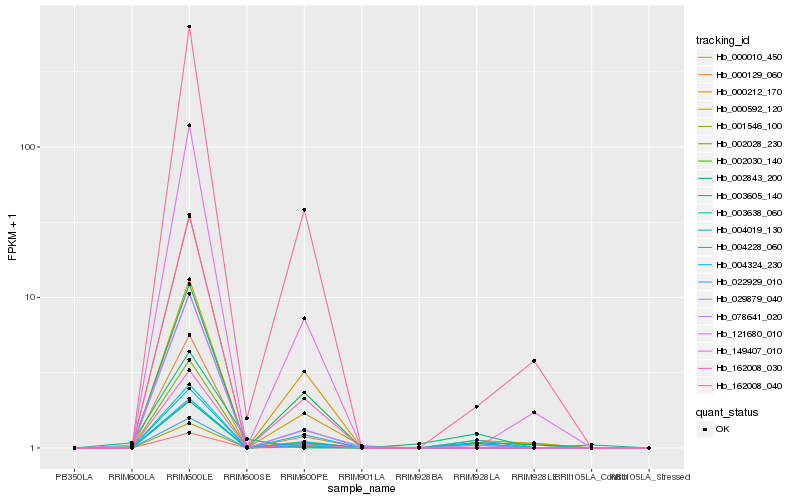
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_450 |
0.0 |
- |
- |
purine permease, putative [Ricinus communis] |
| 2 |
Hb_004324_230 |
0.1375413251 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 3 |
Hb_004228_060 |
0.1749629303 |
- |
- |
Tubulin beta-2 chain [Triticum urartu] |
| 4 |
Hb_003638_060 |
0.178744911 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 5 |
Hb_000592_120 |
0.2360174836 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 1 [Jatropha curcas] |
| 6 |
Hb_003605_140 |
0.248723918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_078641_020 |
0.249509908 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 8 |
Hb_000212_170 |
0.262711067 |
- |
- |
PREDICTED: GDSL esterase/lipase At2g30310-like [Jatropha curcas] |
| 9 |
Hb_002843_200 |
0.2660932311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_162008_040 |
0.26649861 |
- |
- |
hypothetical protein CL1725Contig1_03, partial [Pinus taeda] |
| 11 |
Hb_162008_030 |
0.267728155 |
- |
- |
cytochrome p450 [Ageratina adenophora] |
| 12 |
Hb_000129_060 |
0.2679155806 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 13 |
Hb_004019_130 |
0.2682033507 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 14 |
Hb_149407_010 |
0.2692181522 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 15 |
Hb_029879_040 |
0.2696892264 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 16 |
Hb_002028_230 |
0.2701418494 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 17 |
Hb_001546_100 |
0.2721593718 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 18 |
Hb_121680_010 |
0.2726496993 |
- |
- |
PREDICTED: uncharacterized protein LOC105644052 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002030_140 |
0.2732578684 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 20 |
Hb_022929_010 |
0.2735369188 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |