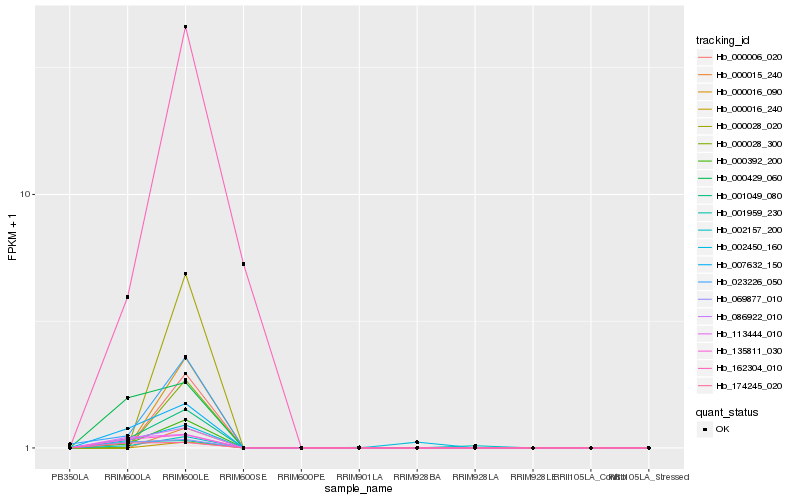
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001049_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_000392_200 |
0.0724117324 |
- |
- |
PREDICTED: cytochrome P450 98A2-like isoform X2 [Phoenix dactylifera] |
| 3 |
Hb_007632_150 |
0.1104321275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_086922_010 |
0.1275214964 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 5 |
Hb_023226_050 |
0.158803326 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 6 |
Hb_135811_030 |
0.2086376112 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 7 |
Hb_069877_010 |
0.2131876368 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 8 |
Hb_000429_060 |
0.2301793821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_113444_010 |
0.2478487404 |
- |
- |
- |
| 10 |
Hb_162304_010 |
0.2518346965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001959_230 |
0.2568225557 |
- |
- |
PREDICTED: uncharacterized protein LOC105638436 [Jatropha curcas] |
| 12 |
Hb_174245_020 |
0.2768071023 |
- |
- |
- |
| 13 |
Hb_002450_160 |
0.2907491079 |
- |
- |
PREDICTED: protein 108 [Jatropha curcas] |
| 14 |
Hb_002157_200 |
0.297881887 |
- |
- |
hypothetical protein VITISV_040170 [Vitis vinifera] |
| 15 |
Hb_000006_020 |
0.3020941477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000015_240 |
0.3020941477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000016_090 |
0.3020941477 |
- |
- |
- |
| 18 |
Hb_000016_240 |
0.3020941477 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0012s13910g [Populus trichocarpa] |
| 19 |
Hb_000028_020 |
0.3020941477 |
- |
- |
- |
| 20 |
Hb_000028_300 |
0.3020941477 |
- |
- |
hypothetical protein 0_13835_01, partial [Pinus radiata] |