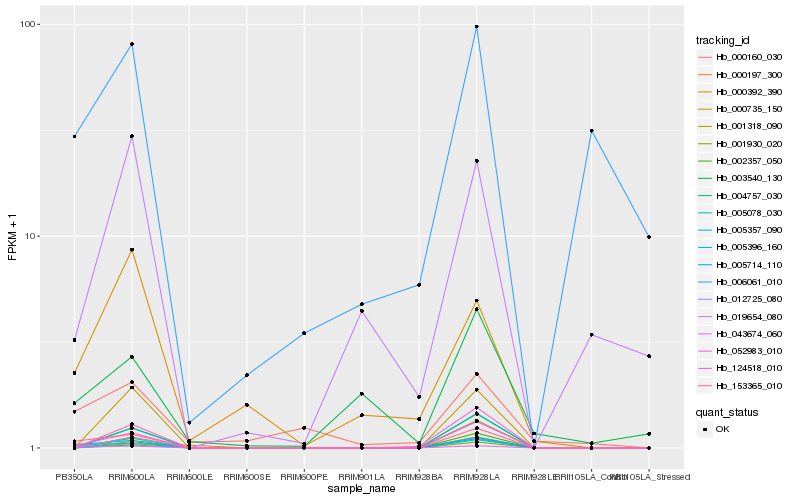
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005714_110 |
0.0 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA10 [Jatropha curcas] |
| 2 |
Hb_153365_010 |
0.0199896193 |
- |
- |
PREDICTED: uncharacterized protein LOC104236681 [Nicotiana sylvestris] |
| 3 |
Hb_043674_060 |
0.0474508498 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 4 |
Hb_052983_010 |
0.2516008311 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_000197_300 |
0.2763088142 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 6 |
Hb_001930_020 |
0.2771586887 |
- |
- |
PREDICTED: pectinesterase QRT1 [Jatropha curcas] |
| 7 |
Hb_005357_090 |
0.2773685332 |
- |
- |
ubiquitin, putative [Ricinus communis] |
| 8 |
Hb_002357_050 |
0.2776950015 |
- |
- |
- |
| 9 |
Hb_124518_010 |
0.2801712577 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 10 |
Hb_005078_030 |
0.2823324264 |
- |
- |
- |
| 11 |
Hb_004757_030 |
0.2826106085 |
- |
- |
- |
| 12 |
Hb_000735_150 |
0.2883366343 |
- |
- |
hypothetical protein JCGZ_15322 [Jatropha curcas] |
| 13 |
Hb_001318_090 |
0.3064104314 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 14 |
Hb_012725_080 |
0.306995811 |
- |
- |
- |
| 15 |
Hb_000392_390 |
0.31057893 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000160_030 |
0.3138338693 |
- |
- |
putative S-adenosylmethionine-dependent methyltransferase [Morus notabilis] |
| 17 |
Hb_005396_160 |
0.3186735183 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 18 |
Hb_003540_130 |
0.3290731063 |
- |
- |
- |
| 19 |
Hb_019654_080 |
0.3309700711 |
- |
- |
- |
| 20 |
Hb_006061_010 |
0.3322016532 |
- |
- |
PREDICTED: phytosulfokines 4-like [Populus euphratica] |