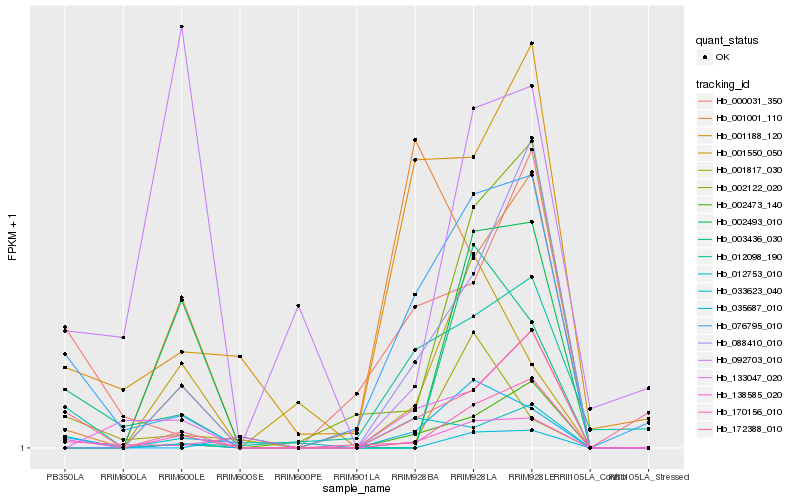
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138585_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_076795_010 |
0.2686314964 |
- |
- |
hypothetical protein POPTR_0001s07680g, partial [Populus trichocarpa] |
| 3 |
Hb_088410_010 |
0.2804513793 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_003436_030 |
0.2933431739 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 5 |
Hb_172388_010 |
0.3006533727 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 6 |
Hb_002122_020 |
0.3020900229 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 7 |
Hb_002493_010 |
0.3054135111 |
- |
- |
- |
| 8 |
Hb_033623_040 |
0.3142545882 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 9 |
Hb_012753_010 |
0.3219505264 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_002473_140 |
0.3236528276 |
- |
- |
- |
| 11 |
Hb_001188_120 |
0.343543381 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 12 |
Hb_000031_350 |
0.3457851777 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic isoform X2 [Cucumis melo] |
| 13 |
Hb_133047_020 |
0.3493427664 |
- |
- |
lysophosphatidic acid acyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001001_110 |
0.351066954 |
- |
- |
- |
| 15 |
Hb_170156_010 |
0.3520827741 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_012098_190 |
0.3566886878 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 17 |
Hb_001817_030 |
0.3589314302 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 18 |
Hb_092703_010 |
0.3593573519 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 19 |
Hb_001550_050 |
0.3601055936 |
- |
- |
PREDICTED: J domain-containing protein spf31 [Eucalyptus grandis] |
| 20 |
Hb_035687_010 |
0.3626741841 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |