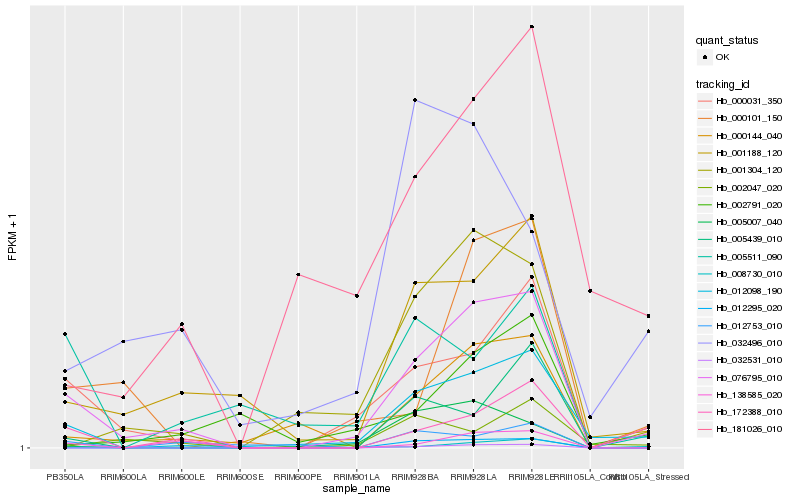
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_076795_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s07680g, partial [Populus trichocarpa] |
| 2 |
Hb_000031_350 |
0.1973602591 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic isoform X2 [Cucumis melo] |
| 3 |
Hb_012098_190 |
0.1987329751 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 4 |
Hb_001188_120 |
0.2344860036 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 5 |
Hb_000144_040 |
0.2528566891 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 6 |
Hb_172388_010 |
0.252984573 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 7 |
Hb_008730_010 |
0.2574210121 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_000101_150 |
0.2631998277 |
- |
- |
- |
| 9 |
Hb_138585_020 |
0.2686314964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001304_120 |
0.2759201437 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 11 |
Hb_005007_040 |
0.3037163731 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 12 |
Hb_002047_020 |
0.3078159724 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_032531_010 |
0.3083042817 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_005511_090 |
0.3092363718 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 C isoform X1 [Jatropha curcas] |
| 15 |
Hb_005439_010 |
0.3104051065 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_032496_010 |
0.3125847744 |
- |
- |
hypothetical protein POPTR_0012s10850g [Populus trichocarpa] |
| 17 |
Hb_012753_010 |
0.3135220821 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_181026_010 |
0.3160723248 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 19 |
Hb_002791_020 |
0.3161227347 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 20 |
Hb_012295_020 |
0.3171038975 |
- |
- |
PREDICTED: uncharacterized protein LOC104223093 [Nicotiana sylvestris] |