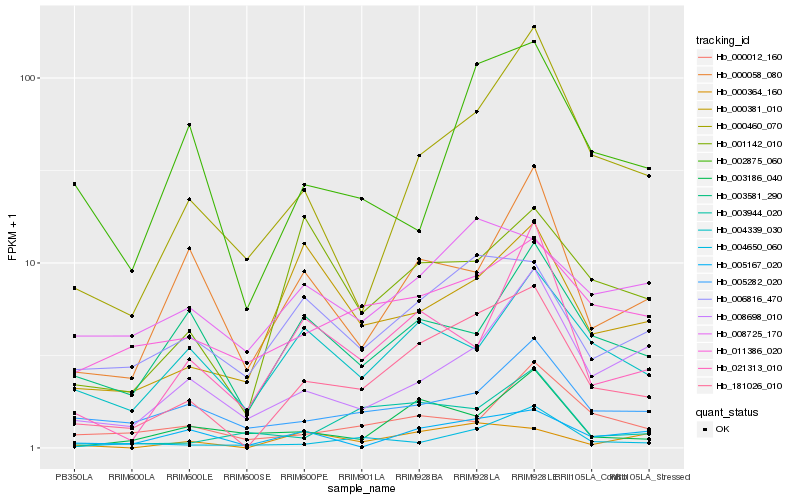
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181026_010 |
0.0 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 2 |
Hb_000460_070 |
0.2058068984 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 3 |
Hb_006816_470 |
0.214348583 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011386_020 |
0.2241667353 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_005167_020 |
0.2273692924 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 6 |
Hb_004650_060 |
0.2292947725 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 7 |
Hb_004339_030 |
0.231192519 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 8 |
Hb_008698_010 |
0.2319563606 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_005282_020 |
0.233141409 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 10 |
Hb_002875_060 |
0.2343457882 |
- |
- |
hypothetical protein L484_023461 [Morus notabilis] |
| 11 |
Hb_000058_080 |
0.2355244443 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_001142_010 |
0.2365364622 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 13 |
Hb_021313_010 |
0.2366643092 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 14 |
Hb_008725_170 |
0.2388053911 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 15 |
Hb_003581_290 |
0.2389388116 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_000364_160 |
0.2390312717 |
- |
- |
PREDICTED: uncharacterized protein LOC105639754 [Jatropha curcas] |
| 17 |
Hb_003186_040 |
0.2406780293 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 18 |
Hb_000012_160 |
0.2440002322 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000381_010 |
0.2440417348 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_003944_020 |
0.2468881897 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |