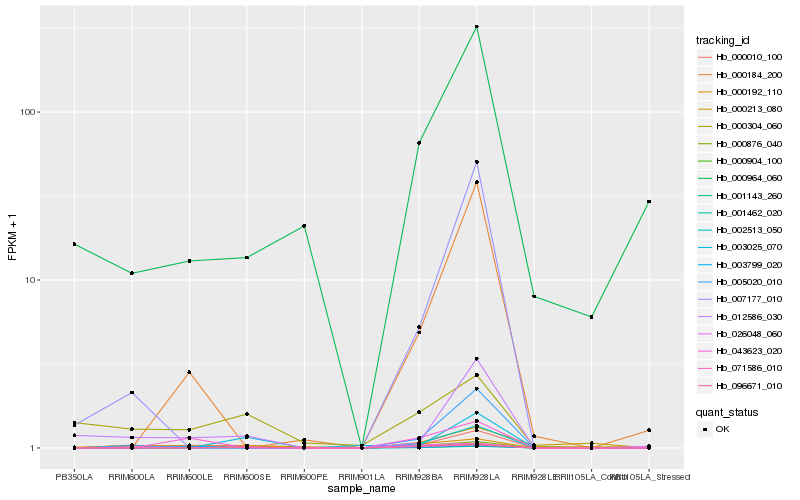
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 2 |
Hb_026048_060 |
0.2337962531 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_012586_030 |
0.246123212 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 4 |
Hb_001143_260 |
0.2853343265 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 5 |
Hb_002513_050 |
0.2863880745 |
- |
- |
hypothetical protein F383_14794 [Gossypium arboreum] |
| 6 |
Hb_000184_200 |
0.301943719 |
- |
- |
- |
| 7 |
Hb_003799_020 |
0.3039404373 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 8 |
Hb_000876_040 |
0.3094515778 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 9 |
Hb_043623_020 |
0.3155173887 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 10 |
Hb_005020_010 |
0.3175648811 |
- |
- |
- |
| 11 |
Hb_000304_060 |
0.3198913951 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 12 |
Hb_000904_100 |
0.3225726338 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 13 |
Hb_096671_010 |
0.3236248966 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 14 |
Hb_001462_020 |
0.3271949829 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 15 |
Hb_071586_010 |
0.3277474225 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 16 |
Hb_007177_010 |
0.3288548979 |
- |
- |
- |
| 17 |
Hb_000964_060 |
0.3314216236 |
- |
- |
hypothetical protein POPTR_0009s06660g [Populus trichocarpa] |
| 18 |
Hb_000192_110 |
0.3352182718 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 19 |
Hb_000213_080 |
0.337167394 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Vitis vinifera] |
| 20 |
Hb_003025_070 |
0.3464237612 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |