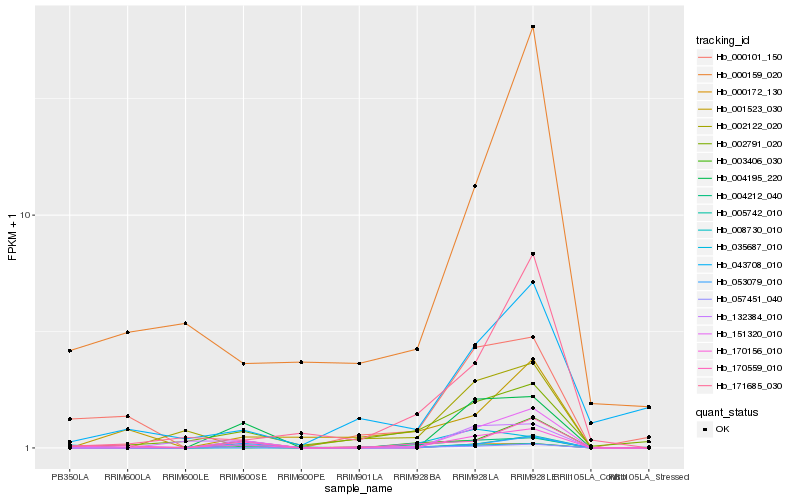
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_170156_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_002791_020 |
0.232061394 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 3 |
Hb_000101_150 |
0.2486498492 |
- |
- |
- |
| 4 |
Hb_170559_010 |
0.249549131 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_000159_020 |
0.264306733 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 6 |
Hb_003406_030 |
0.2672807024 |
- |
- |
PREDICTED: uncharacterized protein LOC104774100 [Camelina sativa] |
| 7 |
Hb_151320_010 |
0.2684592955 |
- |
- |
- |
| 8 |
Hb_004212_040 |
0.2731131195 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_043708_010 |
0.2757122912 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 10 |
Hb_057451_040 |
0.2865444742 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_000172_130 |
0.2927355696 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 12 |
Hb_171685_030 |
0.2934219698 |
- |
- |
URF-1 [Citrullus lanatus] |
| 13 |
Hb_053079_010 |
0.2943130897 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 14 |
Hb_002122_020 |
0.2954007116 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 15 |
Hb_008730_010 |
0.2967490235 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_035687_010 |
0.2997358415 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 17 |
Hb_004195_220 |
0.2998963137 |
- |
- |
hypothetical protein PRUPE_ppa005288mg [Prunus persica] |
| 18 |
Hb_005742_010 |
0.3018990097 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 19 |
Hb_132384_010 |
0.3047735944 |
- |
- |
hypothetical protein VITISV_018559 [Vitis vinifera] |
| 20 |
Hb_001523_030 |
0.3052871827 |
- |
- |
protein binding protein, putative [Ricinus communis] |