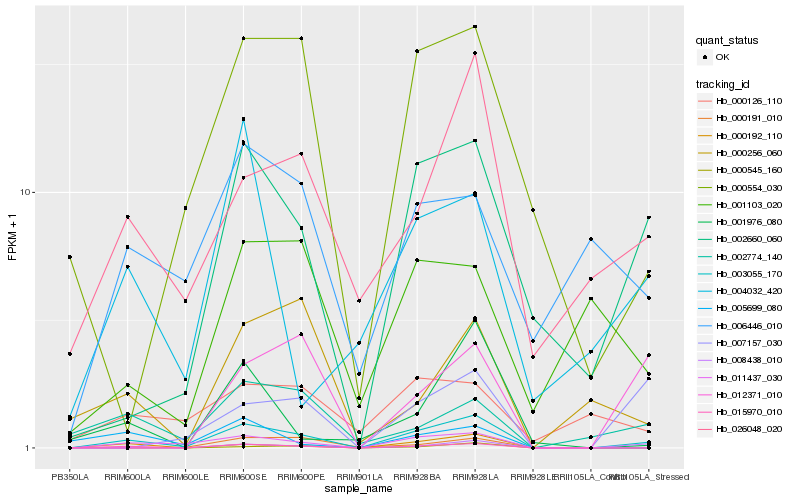
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003055_170 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25 isoform X1 [Jatropha curcas] |
| 2 |
Hb_015970_010 |
0.1950603746 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 3 |
Hb_011437_030 |
0.2213780295 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |
| 4 |
Hb_002660_060 |
0.2606167633 |
- |
- |
PREDICTED: uncharacterized protein LOC105642215 [Jatropha curcas] |
| 5 |
Hb_000192_110 |
0.2728712083 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 6 |
Hb_026048_020 |
0.2857591643 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |
| 7 |
Hb_008438_010 |
0.3071534536 |
- |
- |
PREDICTED: uncharacterized protein LOC104245771 [Nicotiana sylvestris] |
| 8 |
Hb_002774_140 |
0.3072985702 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001976_080 |
0.3130463691 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 10 |
Hb_000554_030 |
0.3130623529 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007157_030 |
0.318592394 |
- |
- |
PREDICTED: protein LURP-one-related 6-like [Jatropha curcas] |
| 12 |
Hb_005699_080 |
0.3294317054 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 13 |
Hb_000256_060 |
0.3311531087 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 14 |
Hb_000545_160 |
0.331237932 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 15 |
Hb_000191_010 |
0.3330724491 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_000126_110 |
0.3337650039 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 17 |
Hb_006446_010 |
0.3358007179 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_004032_420 |
0.3382831855 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 19 |
Hb_012371_010 |
0.3385102796 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 20 |
Hb_001103_020 |
0.3395206789 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |