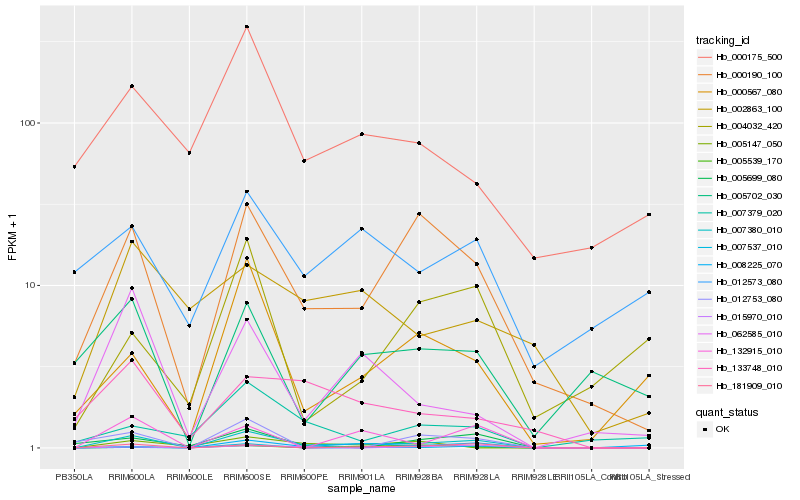
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007380_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_L00213, partial [Eucalyptus grandis] |
| 2 |
Hb_181909_010 |
0.2465027359 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 3 |
Hb_000190_100 |
0.2500063003 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 4 |
Hb_132915_010 |
0.2707137276 |
- |
- |
PREDICTED: uncharacterized protein LOC103700431, partial [Phoenix dactylifera] |
| 5 |
Hb_062585_010 |
0.3137828043 |
- |
- |
hypothetical protein JCGZ_19532 [Jatropha curcas] |
| 6 |
Hb_007537_010 |
0.3144736848 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000567_080 |
0.3152582246 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 8 |
Hb_015970_010 |
0.3205483305 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 9 |
Hb_005699_080 |
0.3293667884 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 10 |
Hb_133748_010 |
0.3330129127 |
- |
- |
- |
| 11 |
Hb_008225_070 |
0.3419335235 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000175_500 |
0.3466306399 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 13 |
Hb_005702_030 |
0.3477863677 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 14 |
Hb_012753_080 |
0.3483435914 |
- |
- |
- |
| 15 |
Hb_007379_020 |
0.3516577186 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 16 |
Hb_002863_100 |
0.3555183321 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 17 |
Hb_005539_170 |
0.3569231898 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 18 |
Hb_012573_080 |
0.3579077945 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 19 |
Hb_005147_050 |
0.3581383599 |
- |
- |
PREDICTED: ankyrin-2-like [Citrus sinensis] |
| 20 |
Hb_004032_420 |
0.3633411472 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |