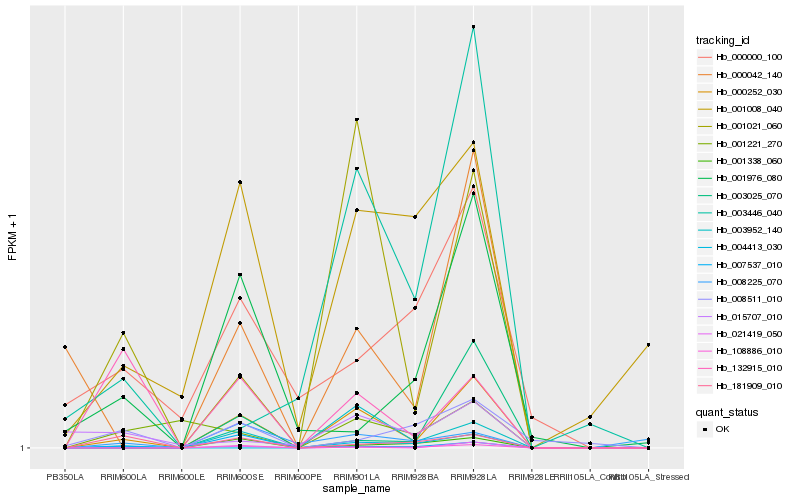
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_030 |
0.0 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 2 |
Hb_108886_010 |
0.2492095682 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001021_060 |
0.2698137741 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003952_140 |
0.2724166093 |
- |
- |
hypothetical protein POPTR_0001s24960g [Populus trichocarpa] |
| 5 |
Hb_021419_050 |
0.2904070284 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Malus domestica] |
| 6 |
Hb_001338_060 |
0.3087952981 |
- |
- |
- |
| 7 |
Hb_000042_140 |
0.31044638 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 8 |
Hb_003025_070 |
0.3216218707 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 9 |
Hb_001008_040 |
0.3331010589 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 10 |
Hb_003446_040 |
0.3397820976 |
- |
- |
- |
| 11 |
Hb_132915_010 |
0.3515645568 |
- |
- |
PREDICTED: uncharacterized protein LOC103700431, partial [Phoenix dactylifera] |
| 12 |
Hb_001976_080 |
0.3518643419 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 13 |
Hb_181909_010 |
0.355745209 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 14 |
Hb_000000_100 |
0.357483915 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 15 |
Hb_001221_270 |
0.3639464136 |
- |
- |
- |
| 16 |
Hb_015707_010 |
0.3684931439 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 17 |
Hb_007537_010 |
0.369399588 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_004413_030 |
0.3715360716 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 19 |
Hb_008225_070 |
0.3795090712 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_008511_010 |
0.3824013908 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK6 [Jatropha curcas] |