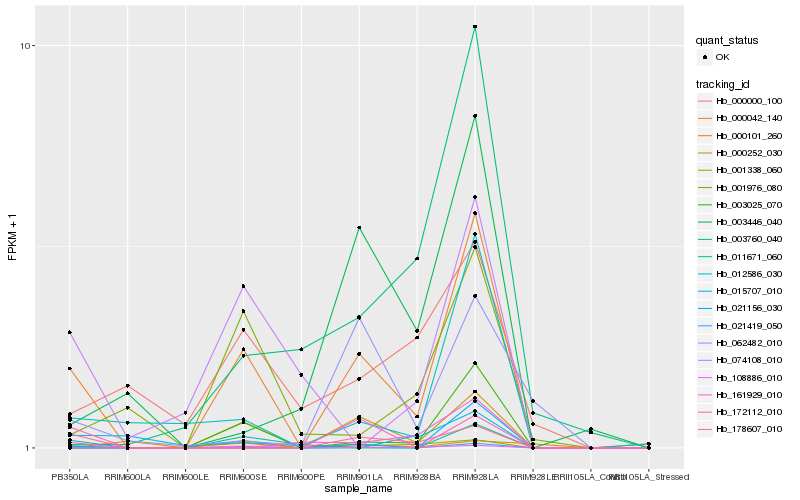
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 2 |
Hb_003025_070 |
0.2974150449 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 3 |
Hb_021419_050 |
0.2979743997 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Malus domestica] |
| 4 |
Hb_108886_010 |
0.3080721351 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000252_030 |
0.31044638 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 6 |
Hb_178607_010 |
0.3418816368 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 7 |
Hb_015707_010 |
0.3582042867 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 8 |
Hb_001338_060 |
0.3595419582 |
- |
- |
- |
| 9 |
Hb_000101_260 |
0.3659636892 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_011671_060 |
0.3677561848 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 11 |
Hb_001976_080 |
0.3704420207 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 12 |
Hb_074108_010 |
0.3720978433 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 13 |
Hb_000000_100 |
0.3754532578 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 14 |
Hb_003446_040 |
0.3767538636 |
- |
- |
- |
| 15 |
Hb_003760_040 |
0.3786062366 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 16 |
Hb_062482_010 |
0.390773644 |
- |
- |
- |
| 17 |
Hb_161929_010 |
0.3914051433 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 18 |
Hb_021156_030 |
0.3916465132 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 19 |
Hb_172112_010 |
0.3946102398 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |
| 20 |
Hb_012586_030 |
0.395666248 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |