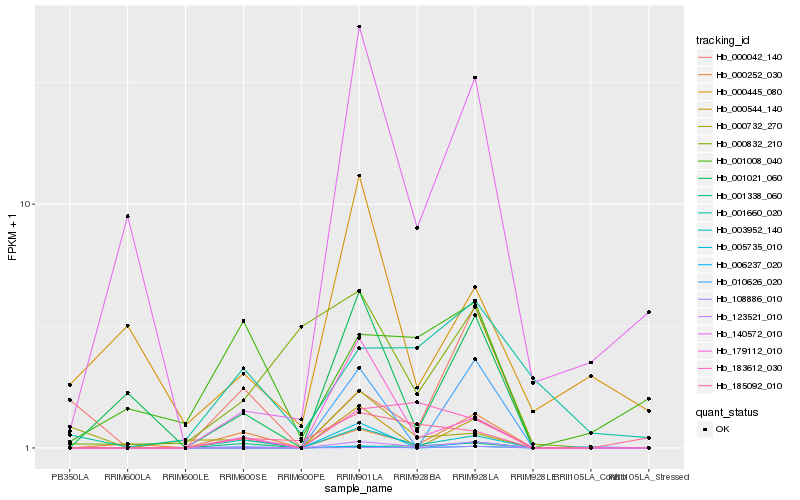
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003952_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s24960g [Populus trichocarpa] |
| 2 |
Hb_000252_030 |
0.2724166093 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 3 |
Hb_179112_010 |
0.2724404249 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 4 |
Hb_001021_060 |
0.2866493771 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_123521_010 |
0.3222046891 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 6 |
Hb_108886_010 |
0.3463211917 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_183612_030 |
0.3516593747 |
- |
- |
PREDICTED: uncharacterized protein LOC105632256 [Jatropha curcas] |
| 8 |
Hb_000544_140 |
0.3649849861 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 9 |
Hb_006237_020 |
0.3680283342 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_001338_060 |
0.3729006191 |
- |
- |
- |
| 11 |
Hb_140572_010 |
0.3736412663 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 12 |
Hb_185092_010 |
0.3795511163 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001008_040 |
0.3818714444 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 14 |
Hb_000832_210 |
0.3845944563 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 15 |
Hb_010626_020 |
0.3880051744 |
- |
- |
- |
| 16 |
Hb_000732_270 |
0.4014509437 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 17 |
Hb_005735_010 |
0.4015695266 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000445_080 |
0.403898906 |
transcription factor |
TF Family: MIKC |
PREDICTED: floral homeotic protein PMADS 2 [Jatropha curcas] |
| 19 |
Hb_000042_140 |
0.4052464394 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 20 |
Hb_001660_020 |
0.4057331566 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |