| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
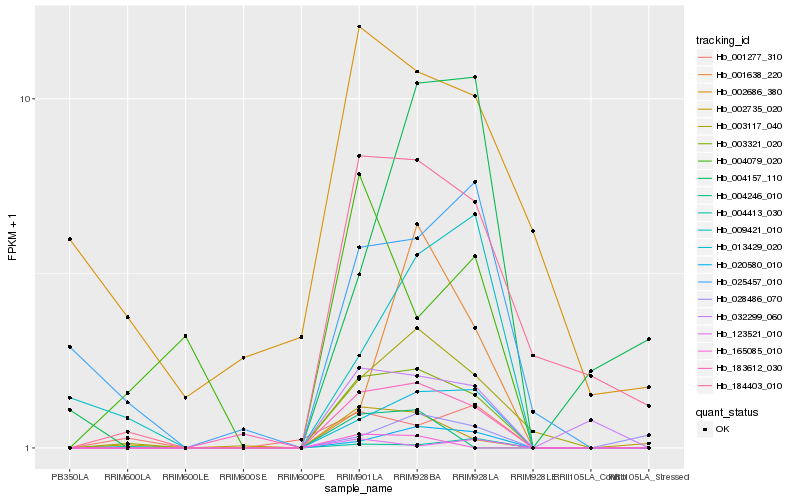
Hb_003321_020 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 2 |
Hb_013429_020 |
0.18807674 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_183612_030 |
0.1896366817 |
- |
- |
PREDICTED: uncharacterized protein LOC105632256 [Jatropha curcas] |
| 4 |
Hb_003117_040 |
0.2010261471 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_020580_010 |
0.2079923461 |
- |
- |
- |
| 6 |
Hb_032299_060 |
0.2346069339 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Elaeis guineensis] |
| 7 |
Hb_184403_010 |
0.2385820664 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Gossypium raimondii] |
| 8 |
Hb_002735_020 |
0.2876405355 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_123521_010 |
0.315234195 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 10 |
Hb_025457_010 |
0.325052212 |
- |
- |
- |
| 11 |
Hb_001277_310 |
0.3341254434 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 12 |
Hb_001638_220 |
0.3353293478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_028486_070 |
0.3376103624 |
- |
- |
hypothetical protein CISIN_1g028119mg [Citrus sinensis] |
| 14 |
Hb_009421_010 |
0.3426263498 |
- |
- |
- |
| 15 |
Hb_004157_110 |
0.3460980008 |
- |
- |
- |
| 16 |
Hb_004413_030 |
0.3563450252 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 17 |
Hb_004079_020 |
0.3654800955 |
- |
- |
hypothetical protein glysoja_031528 [Glycine soja] |
| 18 |
Hb_002686_380 |
0.3656009779 |
- |
- |
- |
| 19 |
Hb_004246_010 |
0.3683722418 |
- |
- |
- |
| 20 |
Hb_165085_010 |
0.3715558359 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |