| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
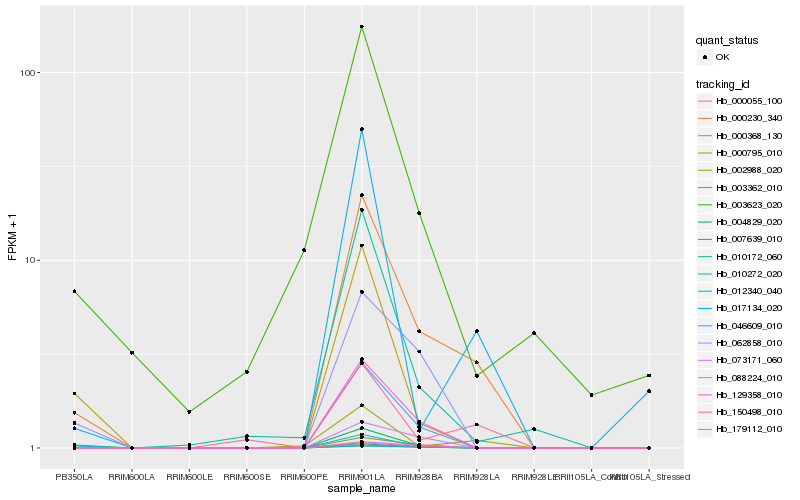
Hb_000230_340 |
0.0 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 2 |
Hb_046609_010 |
0.2151963012 |
- |
- |
- |
| 3 |
Hb_129358_010 |
0.2184820621 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 11-like [Jatropha curcas] |
| 4 |
Hb_007639_010 |
0.2245244737 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 5 |
Hb_002988_020 |
0.2298206563 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 6 |
Hb_179112_010 |
0.2351477477 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 7 |
Hb_003362_010 |
0.2364475784 |
- |
- |
Actin-related protein 2/3 complex subunit 4 [Morus notabilis] |
| 8 |
Hb_150498_010 |
0.2430452008 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 9 |
Hb_004829_020 |
0.2453693537 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_010172_060 |
0.2474494955 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 11 |
Hb_062858_010 |
0.2490278185 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 12 |
Hb_012340_040 |
0.2495851569 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase-like [Populus euphratica] |
| 13 |
Hb_017134_020 |
0.252892666 |
- |
- |
- |
| 14 |
Hb_010272_020 |
0.2569143365 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 15 |
Hb_073171_060 |
0.2631954411 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 16 |
Hb_000795_010 |
0.2660135644 |
- |
- |
- |
| 17 |
Hb_003623_020 |
0.2670885269 |
- |
- |
- |
| 18 |
Hb_000055_100 |
0.2704669596 |
- |
- |
polyprotein [Oryza australiensis] |
| 19 |
Hb_000368_130 |
0.270914588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_088224_010 |
0.2736801053 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |