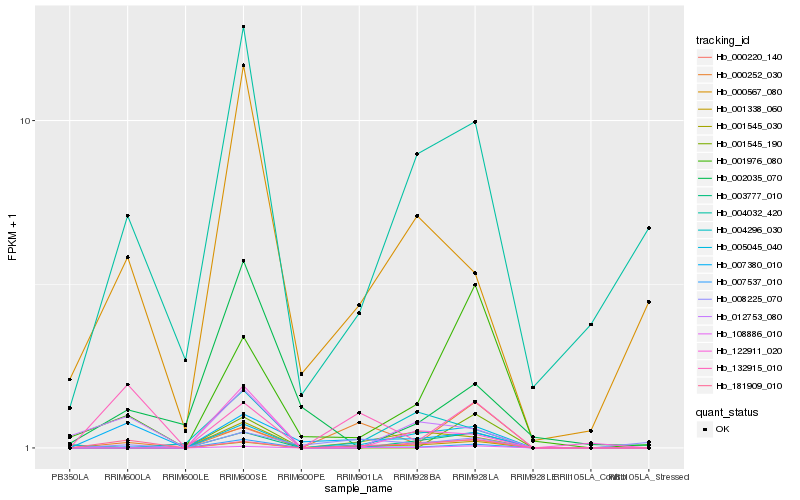
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007537_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_108886_010 |
0.3060850501 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001338_060 |
0.3066941429 |
- |
- |
- |
| 4 |
Hb_007380_010 |
0.3144736848 |
- |
- |
hypothetical protein EUGRSUZ_L00213, partial [Eucalyptus grandis] |
| 5 |
Hb_005045_040 |
0.3155546742 |
- |
- |
hypothetical protein POPTR_0017s04090g, partial [Populus trichocarpa] |
| 6 |
Hb_000567_080 |
0.3320148387 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 7 |
Hb_001976_080 |
0.3325208027 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 8 |
Hb_004032_420 |
0.3357288724 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 9 |
Hb_181909_010 |
0.3394863126 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 10 |
Hb_003777_010 |
0.3435813464 |
- |
- |
- |
| 11 |
Hb_002035_070 |
0.3466831583 |
- |
- |
hypothetical protein CICLE_v10033569mg [Citrus clementina] |
| 12 |
Hb_122911_020 |
0.3565298544 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_001545_030 |
0.360188599 |
- |
- |
- |
| 14 |
Hb_008225_070 |
0.3606082711 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_000220_140 |
0.363913454 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 16 |
Hb_001545_190 |
0.3659246165 |
- |
- |
hypothetical protein VITISV_012031 [Vitis vinifera] |
| 17 |
Hb_000252_030 |
0.369399588 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 18 |
Hb_012753_080 |
0.3742812167 |
- |
- |
- |
| 19 |
Hb_004296_030 |
0.3764154727 |
- |
- |
PREDICTED: gamma-tubulin complex component 3 [Jatropha curcas] |
| 20 |
Hb_132915_010 |
0.3789313509 |
- |
- |
PREDICTED: uncharacterized protein LOC103700431, partial [Phoenix dactylifera] |