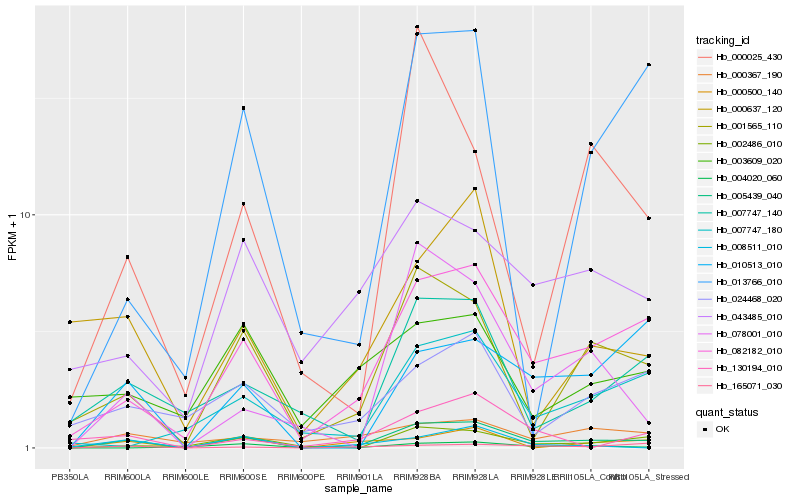
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005439_040 |
0.0 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 2 |
Hb_082182_010 |
0.1580935658 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 3 |
Hb_001565_110 |
0.2228246771 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 4 |
Hb_002486_010 |
0.2389135708 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 1 [Jatropha curcas] |
| 5 |
Hb_043485_010 |
0.2623581402 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000500_140 |
0.2651117071 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 7 |
Hb_007747_140 |
0.2661440396 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_078001_010 |
0.2662787496 |
- |
- |
Uncharacterized protein TCM_000398 [Theobroma cacao] |
| 9 |
Hb_010513_010 |
0.2699878387 |
- |
- |
- |
| 10 |
Hb_007747_180 |
0.2740011134 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |
| 11 |
Hb_013766_010 |
0.2808840073 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 12 |
Hb_165071_030 |
0.280912122 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_000367_190 |
0.2822827729 |
- |
- |
hypothetical protein RCOM_1470550 [Ricinus communis] |
| 14 |
Hb_024468_020 |
0.283608952 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 15 |
Hb_008511_010 |
0.2841456709 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK6 [Jatropha curcas] |
| 16 |
Hb_000637_120 |
0.2926356083 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 17 |
Hb_130194_010 |
0.2944672738 |
- |
- |
PREDICTED: uncharacterized protein LOC104115665 [Nicotiana tomentosiformis] |
| 18 |
Hb_004020_060 |
0.2961985378 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_003609_020 |
0.2970246046 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 20 |
Hb_000025_430 |
0.299537378 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |