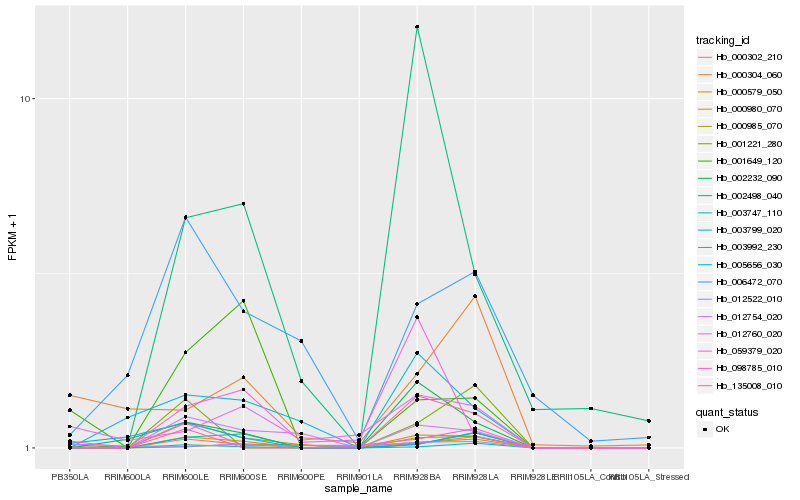
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_050 |
0.0 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 2 |
Hb_000985_070 |
0.3129143773 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 3 |
Hb_003799_020 |
0.3129467155 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 4 |
Hb_002232_090 |
0.3182880033 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 5 |
Hb_000302_210 |
0.3186784121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012760_020 |
0.3226931587 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 7 |
Hb_003747_110 |
0.3351276211 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 8 |
Hb_001649_120 |
0.3407951882 |
- |
- |
- |
| 9 |
Hb_012754_020 |
0.3455514515 |
- |
- |
PREDICTED: uncharacterized protein LOC104229837 [Nicotiana sylvestris] |
| 10 |
Hb_000304_060 |
0.3486835544 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 11 |
Hb_012522_010 |
0.3668371865 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 12 |
Hb_098785_010 |
0.3695014794 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 13 |
Hb_135008_010 |
0.3736817473 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 14 |
Hb_002498_040 |
0.3800663364 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 15 |
Hb_006472_070 |
0.3818445323 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 16 |
Hb_003992_230 |
0.3845386845 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 17 |
Hb_000980_070 |
0.3867940018 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 18 |
Hb_001221_280 |
0.3898666738 |
- |
- |
- |
| 19 |
Hb_059379_020 |
0.3918078057 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 20 |
Hb_005656_030 |
0.3918622507 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |