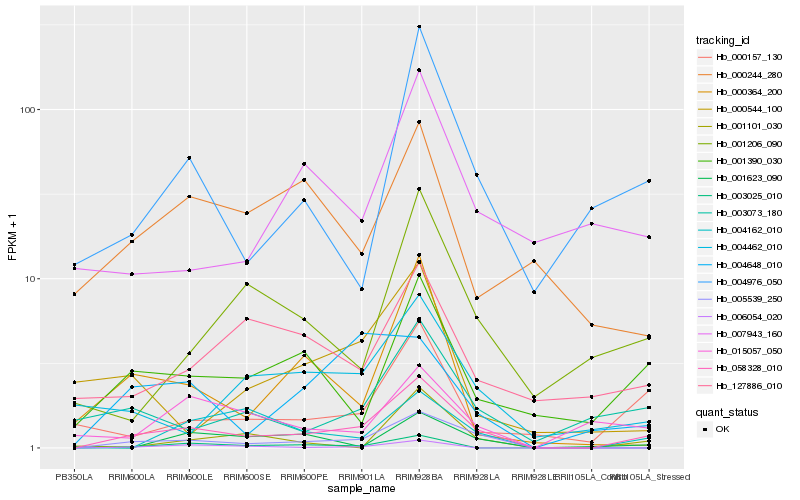
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058328_010 |
0.0 |
- |
- |
JHL06B08.2 [Jatropha curcas] |
| 2 |
Hb_005539_250 |
0.1902308789 |
- |
- |
- |
| 3 |
Hb_001390_030 |
0.2736435468 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 4 |
Hb_000157_130 |
0.277047322 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 5 |
Hb_004976_050 |
0.2780486157 |
- |
- |
- |
| 6 |
Hb_003073_180 |
0.2796105987 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 7 |
Hb_004462_010 |
0.2859158611 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 8 |
Hb_001206_090 |
0.2937420853 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 9 |
Hb_004162_010 |
0.3015617142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000364_200 |
0.3015654449 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003025_010 |
0.3020450742 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 12 |
Hb_127886_010 |
0.30251484 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 13 |
Hb_004648_010 |
0.3050588086 |
- |
- |
UDP-galactose/UDP-glucose transporter 4 [Morus notabilis] |
| 14 |
Hb_015057_050 |
0.3073811057 |
- |
- |
D-alanine aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_001101_030 |
0.3093774384 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 16 |
Hb_006054_020 |
0.3121450532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007943_160 |
0.3129024003 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_000544_100 |
0.3188089946 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 19 |
Hb_001623_090 |
0.3198719037 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 20 |
Hb_000244_280 |
0.3226669428 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |