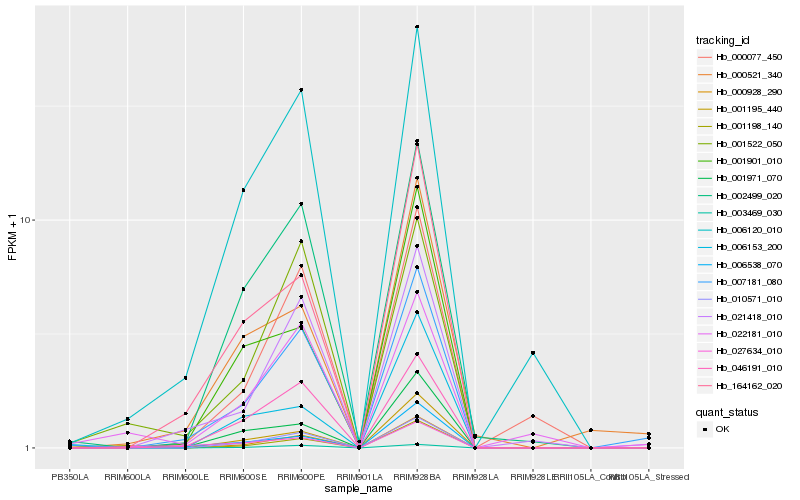
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 2 |
Hb_001198_140 |
0.0515570578 |
- |
- |
- |
| 3 |
Hb_010571_010 |
0.1472452077 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 4 |
Hb_001971_070 |
0.1512740004 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_001522_050 |
0.1675951053 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 6 |
Hb_027634_010 |
0.176349497 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_006120_010 |
0.1804012551 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 8 |
Hb_002499_020 |
0.1809358777 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 9 |
Hb_022181_010 |
0.1818515033 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 10 |
Hb_007181_080 |
0.1835354784 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_164162_020 |
0.1839281478 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 12 |
Hb_001195_440 |
0.183943867 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000077_450 |
0.1864588799 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_006538_070 |
0.188962428 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 15 |
Hb_001901_010 |
0.1898951673 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 16 |
Hb_046191_010 |
0.1900240568 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_003469_030 |
0.1919936208 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 18 |
Hb_021418_010 |
0.1954063051 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 19 |
Hb_006153_200 |
0.1980800046 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Theobroma cacao] |
| 20 |
Hb_000521_340 |
0.2029354789 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |