| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
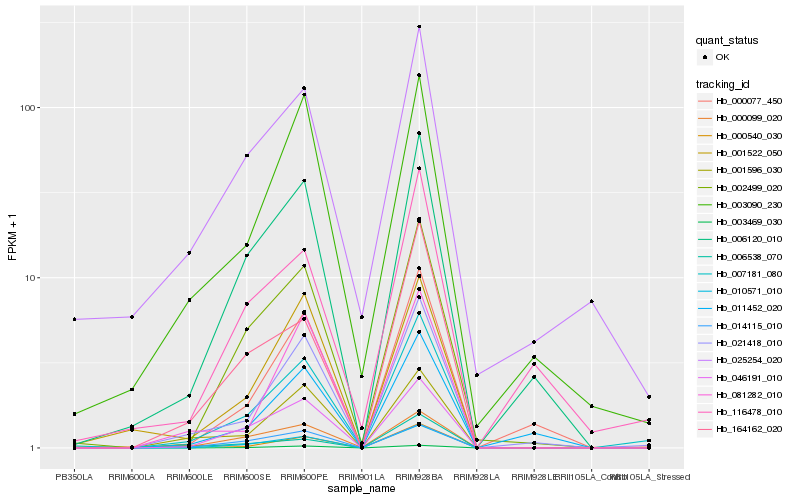
Hb_007181_080 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_021418_010 |
0.1108548967 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 3 |
Hb_010571_010 |
0.1234721913 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 4 |
Hb_002499_020 |
0.1280203086 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 5 |
Hb_046191_010 |
0.1306130146 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_006120_010 |
0.1424298149 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 7 |
Hb_000099_020 |
0.1476137075 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |
| 8 |
Hb_001596_030 |
0.1537713135 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 9 |
Hb_003469_030 |
0.1538760427 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 10 |
Hb_000077_450 |
0.1547293931 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_164162_020 |
0.1550711696 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 12 |
Hb_014115_010 |
0.1568998062 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_003090_230 |
0.1597175423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006538_070 |
0.1600995966 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 15 |
Hb_001522_050 |
0.1624818083 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 16 |
Hb_000540_030 |
0.1628898785 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_081282_010 |
0.1640347535 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 18 |
Hb_116478_010 |
0.1662937868 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 19 |
Hb_025254_020 |
0.1663002385 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 20 |
Hb_011452_020 |
0.1677047081 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |