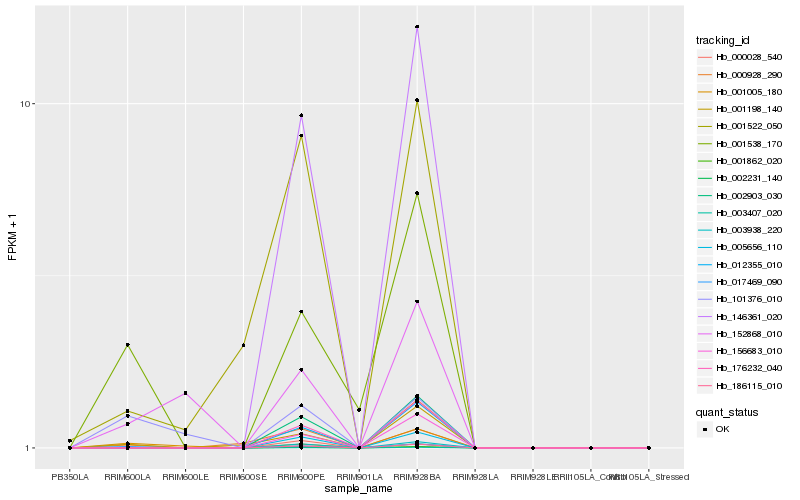
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017469_090 |
0.0 |
- |
- |
Integrase, catalytic region, Zinc finger, CCHC-type, Peptidase aspartic, catalytic, putative [Theobroma cacao] |
| 2 |
Hb_001538_170 |
0.1677681736 |
- |
- |
- |
| 3 |
Hb_001005_180 |
0.2362216749 |
- |
- |
hypothetical protein Csa_5G464830 [Cucumis sativus] |
| 4 |
Hb_101376_010 |
0.2398910962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001198_140 |
0.2464845904 |
- |
- |
- |
| 6 |
Hb_000928_290 |
0.275032838 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 7 |
Hb_152868_010 |
0.3194236191 |
rubber biosynthesis |
Gene Name: REF5 |
small rubber particle protein [Hevea brasiliensis] |
| 8 |
Hb_001862_020 |
0.3195678768 |
- |
- |
PREDICTED: uncharacterized protein LOC103652604 [Zea mays] |
| 9 |
Hb_003407_020 |
0.3196399931 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 10 |
Hb_146361_020 |
0.3196722495 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 11 [Jatropha curcas] |
| 11 |
Hb_186115_010 |
0.3197184538 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_002903_030 |
0.3201985802 |
- |
- |
polyphenol oxidase [Cydonia oblonga] |
| 13 |
Hb_176232_040 |
0.3203917134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012355_010 |
0.3215514138 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 15 |
Hb_003938_220 |
0.3228056125 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_001522_050 |
0.3232469251 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 17 |
Hb_156683_010 |
0.3238388017 |
- |
- |
PREDICTED: uncharacterized protein At5g65660-like [Jatropha curcas] |
| 18 |
Hb_000028_540 |
0.3241466823 |
- |
- |
hypothetical protein POPTR_0012s03720g [Populus trichocarpa] |
| 19 |
Hb_002231_140 |
0.324491866 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 20 |
Hb_005656_110 |
0.3249922655 |
- |
- |
cytochrome P450, putative [Ricinus communis] |