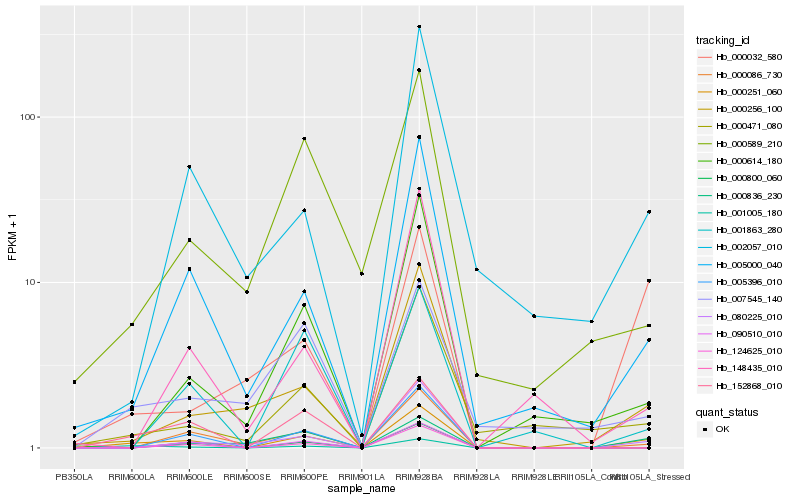
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005000_040 |
0.2275962352 |
- |
- |
ACC oxidase ACCO2 [Manihot esculenta] |
| 3 |
Hb_000086_730 |
0.2445016299 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1-like [Jatropha curcas] |
| 4 |
Hb_080225_010 |
0.244623656 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 5 |
Hb_152868_010 |
0.2566225896 |
rubber biosynthesis |
Gene Name: REF5 |
small rubber particle protein [Hevea brasiliensis] |
| 6 |
Hb_000256_100 |
0.2628161162 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 7 |
Hb_001005_180 |
0.2639281231 |
- |
- |
hypothetical protein Csa_5G464830 [Cucumis sativus] |
| 8 |
Hb_000614_180 |
0.2725337639 |
- |
- |
- |
| 9 |
Hb_001863_280 |
0.2735012588 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 10 |
Hb_000471_080 |
0.2743842158 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 11 |
Hb_000836_230 |
0.2751970563 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 12 |
Hb_000032_580 |
0.275950969 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 13 |
Hb_002057_010 |
0.2765329029 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_005396_010 |
0.2852774688 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 15 |
Hb_124625_010 |
0.2871657757 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 16 |
Hb_090510_010 |
0.2883784138 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 17 |
Hb_000800_060 |
0.2885983092 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 18 |
Hb_148435_010 |
0.2913677552 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 19 |
Hb_007545_140 |
0.2914287574 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 20 |
Hb_000589_210 |
0.2931080541 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |